To determine whether gene transfer from an organelle genome to the nucleus can be observed in the
Question:
a. Leaves from the spectinomycin-resistant transgenic plants were placed in a plant-regeneration medium containing kanamycin. Some of the leaf cells were resistant to kanamycin, and grew into kanamycin-resistant plants. Pollen (paternal) from kanamycin-resistant plants was used to pollinate wild-type (nontransgenic) plants. In tobacco, no chloroplasts are inherited from pollen. The resulting seeds were germinated on media with and without kanamycin. Half of the resulting seedlings were kanamycin resistant. When these kanamycin-resistant plants were allowed to self-pollinate, the offspring exhibited a 3:1 ratio of kanamycin-resistant to sensitive phenotypes. What can be deduced from these data about the location of the kanamycin-resistance gene?
b. To determine whether transfer of the kanamycin-resistance gene to the nucleus was mediated via DNA or an RNA intermediate, DNA was extracted from 10 seedling plants germinated from seeds produced by a wild-type plant pollinated with a kanamycin-resistant plant. The 10 seedling plants, numbered 1-1 0 in the corresponding gel lanes in the figure below, consist of 5 kanamycin-resistant (+) and 5 kanamycin sensitive (-) plants. Each DNA sample was subjected to PCR analysis using primers to amplify the kanamycin-resistance gene (gel at left) or the spectinomycin-resistance gene (gel at right). The lane marked M shows molecular weight markers. What does the correspondence between the presence or absence of PCR products generated in the same plant with both sets of primers suggest about the mode of transfer of the kanamycin gene to the nucleus?
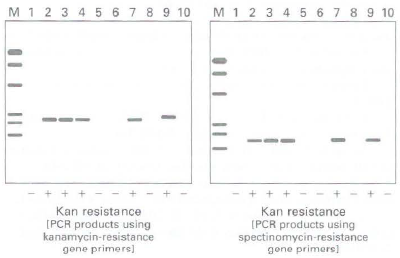
c. When the original transgenic plants, which were selected on spectinomycin but not on kanamycin, were used to pollinate wild-type plants, none of the offspring were kanamycin resistant. What can be deduced from these observations?
Step by Step Answer:

Molecular Cell Biology
ISBN: 978-1429234139
7th edition
Authors: Harvey Lodish, Arnold Berk, Chris A. Kaiser, Monty Krieger, Anthony Bretscher, Hidde Ploegh, Angelika Amon, Matthew P. Scott





