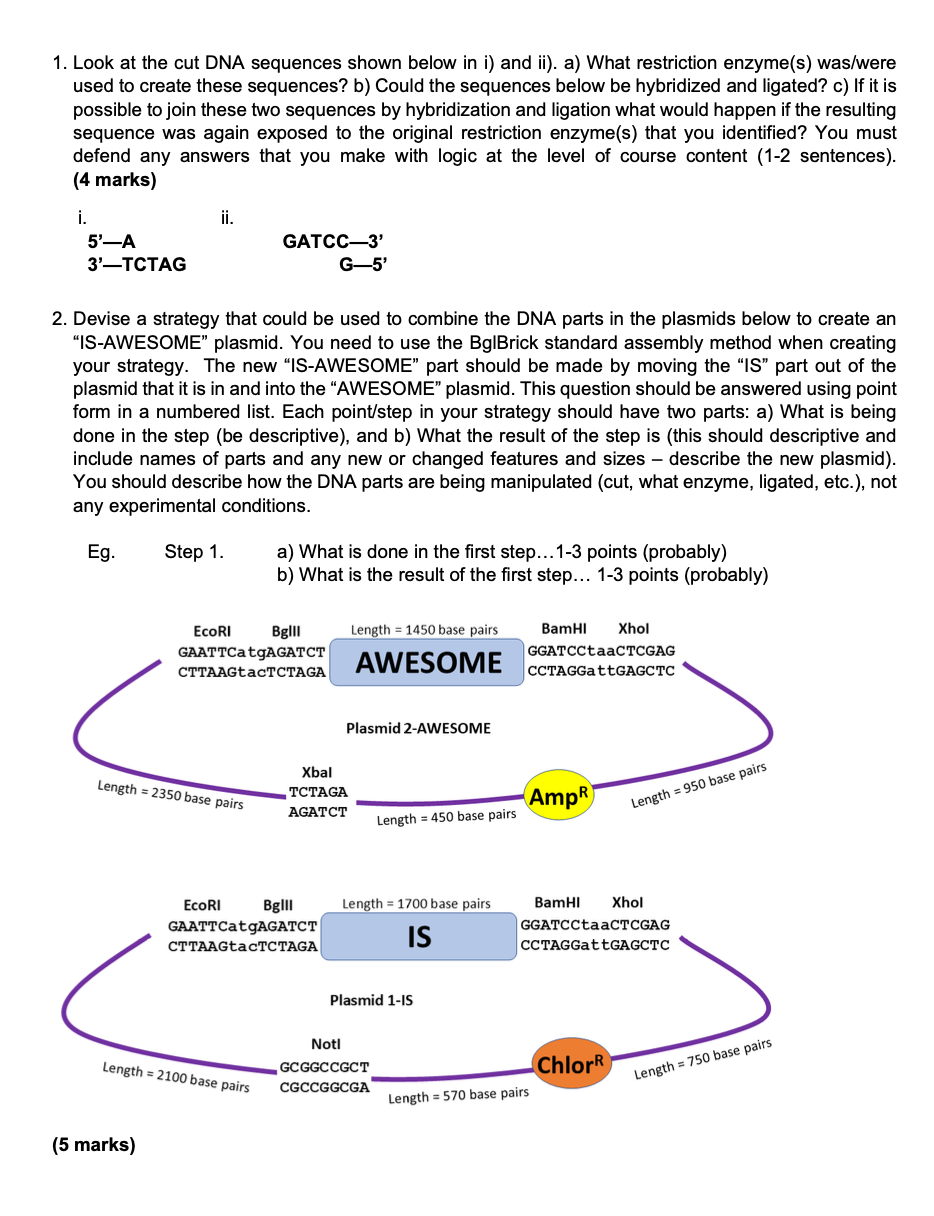
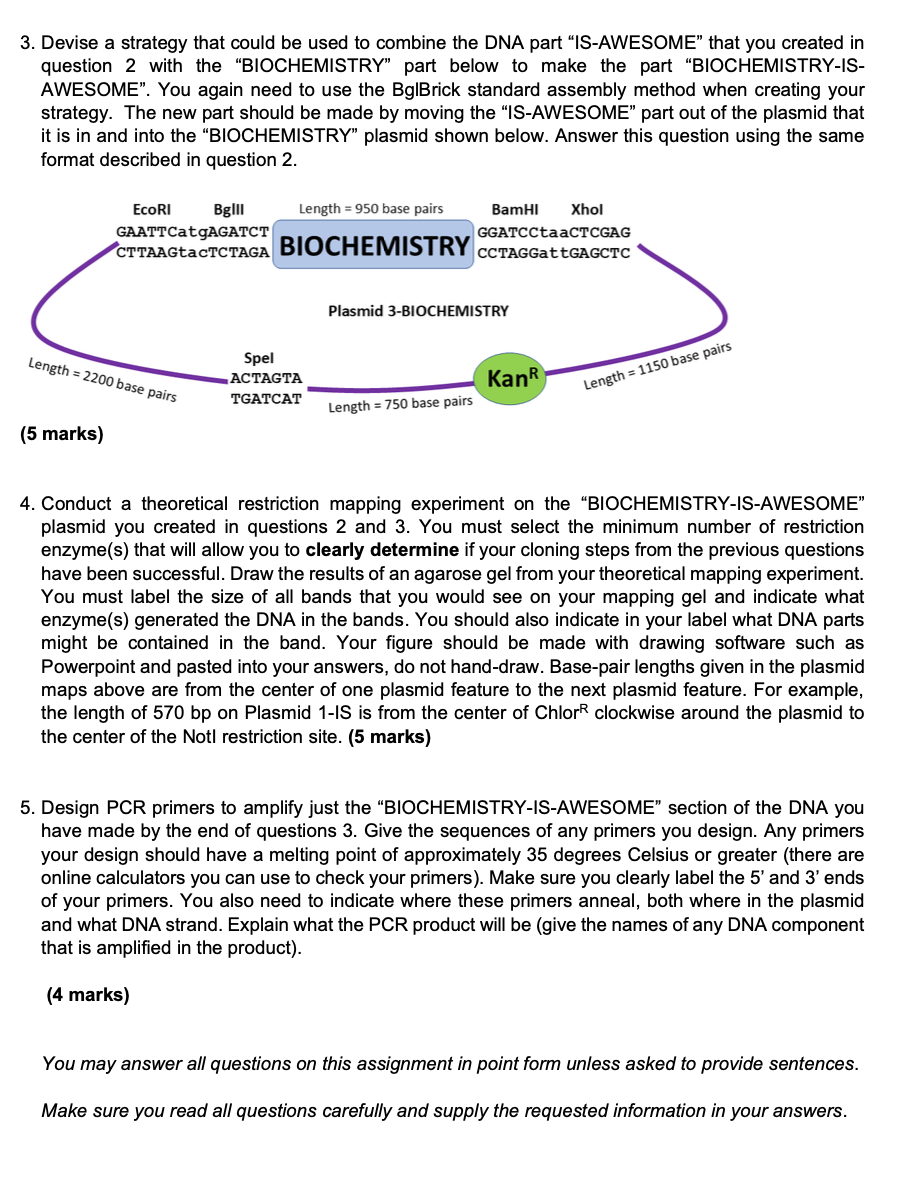
Question: 1. Look at the cut DNA sequences shown below in i) and ii). a) What restriction enzyme(s) was/were used to create these sequences? b) Could
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


