Question: 10 points Save Answer Consider the pseudo code of the motif search using greedy approach discussed in class (in Book it is in section 5.5).
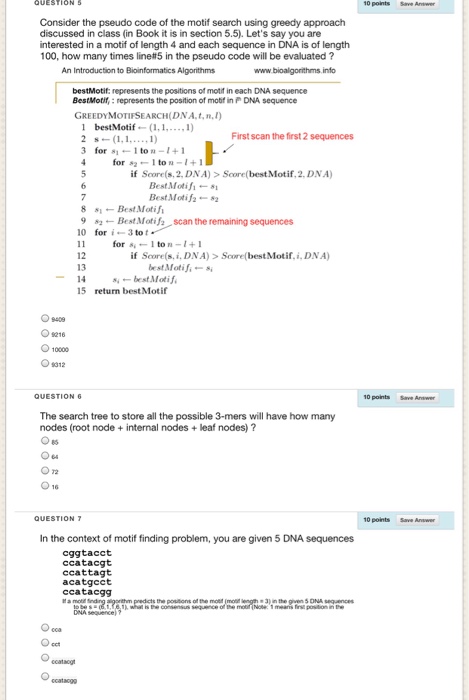
10 points Save Answer Consider the pseudo code of the motif search using greedy approach discussed in class (in Book it is in section 5.5). Let's say you are interested in a motif of length 4 and each sequence in DNA is of length 100, how many times line#5 in the pseudo code will be evaluated ? www.bioalgorithms info bestMotif: represents the positions of motif in each DNA sequence BestMotif,: represents the position of mobf in P DNA sequence GREEDYMOTIFSEARCH(DNA.t,n) 1 bestMotif (. ) 2 s?(1,1, 1) 3 for -1 to u-1+ First scan the first 2 sequences if Soore(s,2, DNA)> Score(best Motif,2. DNA) Best Moti-s Best Motif 82 8 sBestMotifi 92 Best Motif scan the remaining sequences 10 for i-3 tot- for 1 ton +1 if Score(s,i, DNA) > Score(best Motif,i, DNA) 12 13 best Motif-s -14 s. ? bestMotif. 15 return best Motif ? 10000 10 points The search tree to store all the possible 3-mers will have how many nodes (root node internal nodes +leaf nodes) ? 10 points Save Answer In the context of motif finding problem, you are given 5 DNA sequences cggtacct ccatacgt ccattagt acatgcct ccatacgg in the gven 5 ONA sequences
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


