Question: Algorithms Question, thanks Recall the Edit Distance (Sequence Alignment) problem: given two strings over the same alphabet and mismatch and gap penalties, find an alignment
Algorithms Question, thanks
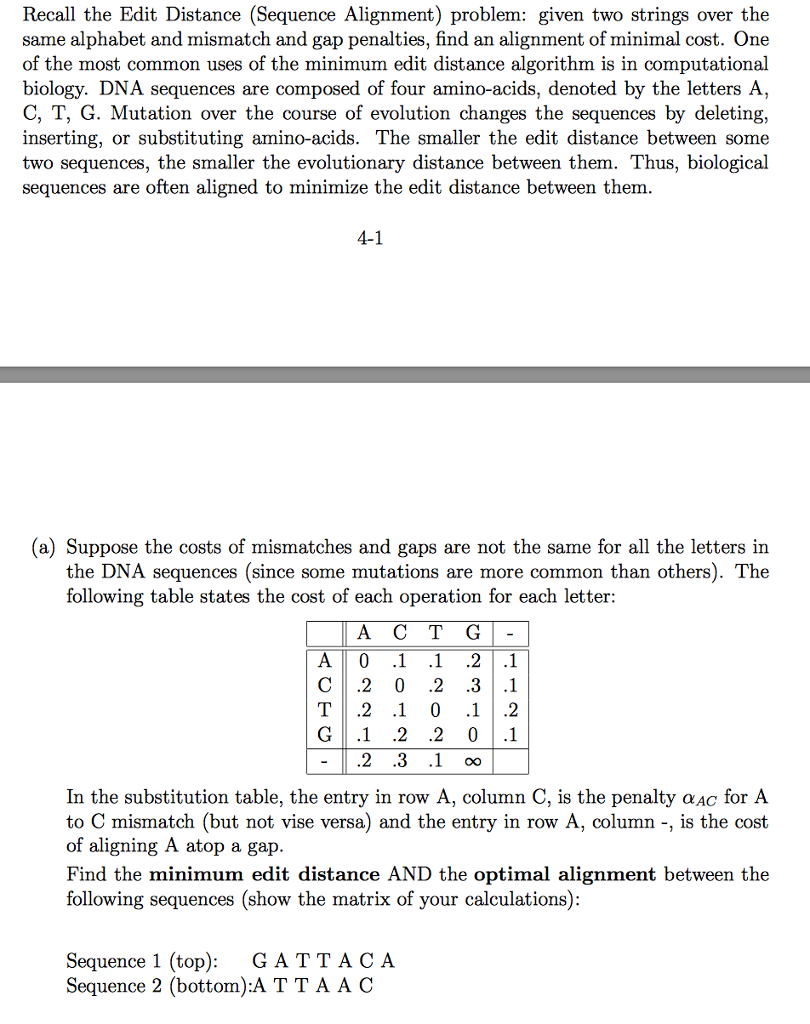
Recall the Edit Distance (Sequence Alignment) problem: given two strings over the same alphabet and mismatch and gap penalties, find an alignment of minimal cost. One of the most common uses of the minimum edit distance algorithm is in computational biology. DNA sequences are composed of four amino-acids, denoted by the letters A, C, T, G. Mutation over the course of evolution changes the sequences by deleting, inserting, or substituting amino-acids. The smaller the edit distance between some two sequences, the smaller the evolutionary distance between them. Thus, biological sequences are often aligned to minimize the edit distance between them 4-1 (a) Suppose the costs of mismatches and gaps are not the same for all the letters in the DNA sequences (since some mutations are more common than others). The following table states the cost of each operation for each letter A C T G C.2 0 .2 .3.1 G1 .2 .2 0 .1 In the substitution table, the entry in row A, column C, is the penalty OAC for A to C mismatch (but not vise versa) and the entry in row A, column -, is the cost of aligning A atop a gap Find the minimum edit distance AND the optimal alignment between the following sequences (show the matrix of your calculations) Sequence 1 (top): GATTACA Sequence 2 (bottom):A TTA A C Recall the Edit Distance (Sequence Alignment) problem: given two strings over the same alphabet and mismatch and gap penalties, find an alignment of minimal cost. One of the most common uses of the minimum edit distance algorithm is in computational biology. DNA sequences are composed of four amino-acids, denoted by the letters A, C, T, G. Mutation over the course of evolution changes the sequences by deleting, inserting, or substituting amino-acids. The smaller the edit distance between some two sequences, the smaller the evolutionary distance between them. Thus, biological sequences are often aligned to minimize the edit distance between them 4-1 (a) Suppose the costs of mismatches and gaps are not the same for all the letters in the DNA sequences (since some mutations are more common than others). The following table states the cost of each operation for each letter A C T G C.2 0 .2 .3.1 G1 .2 .2 0 .1 In the substitution table, the entry in row A, column C, is the penalty OAC for A to C mismatch (but not vise versa) and the entry in row A, column -, is the cost of aligning A atop a gap Find the minimum edit distance AND the optimal alignment between the following sequences (show the matrix of your calculations) Sequence 1 (top): GATTACA Sequence 2 (bottom):A TTA A C
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


