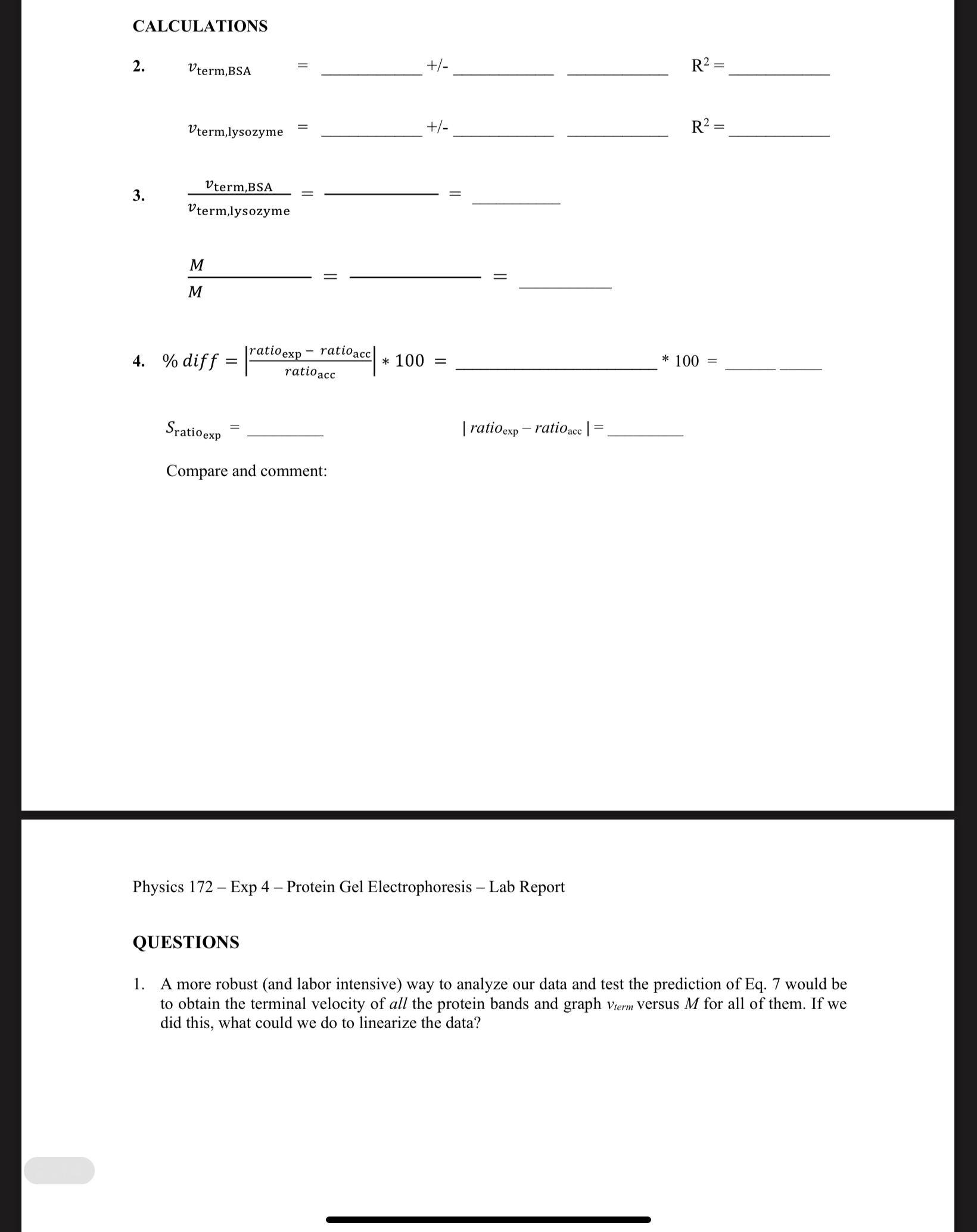
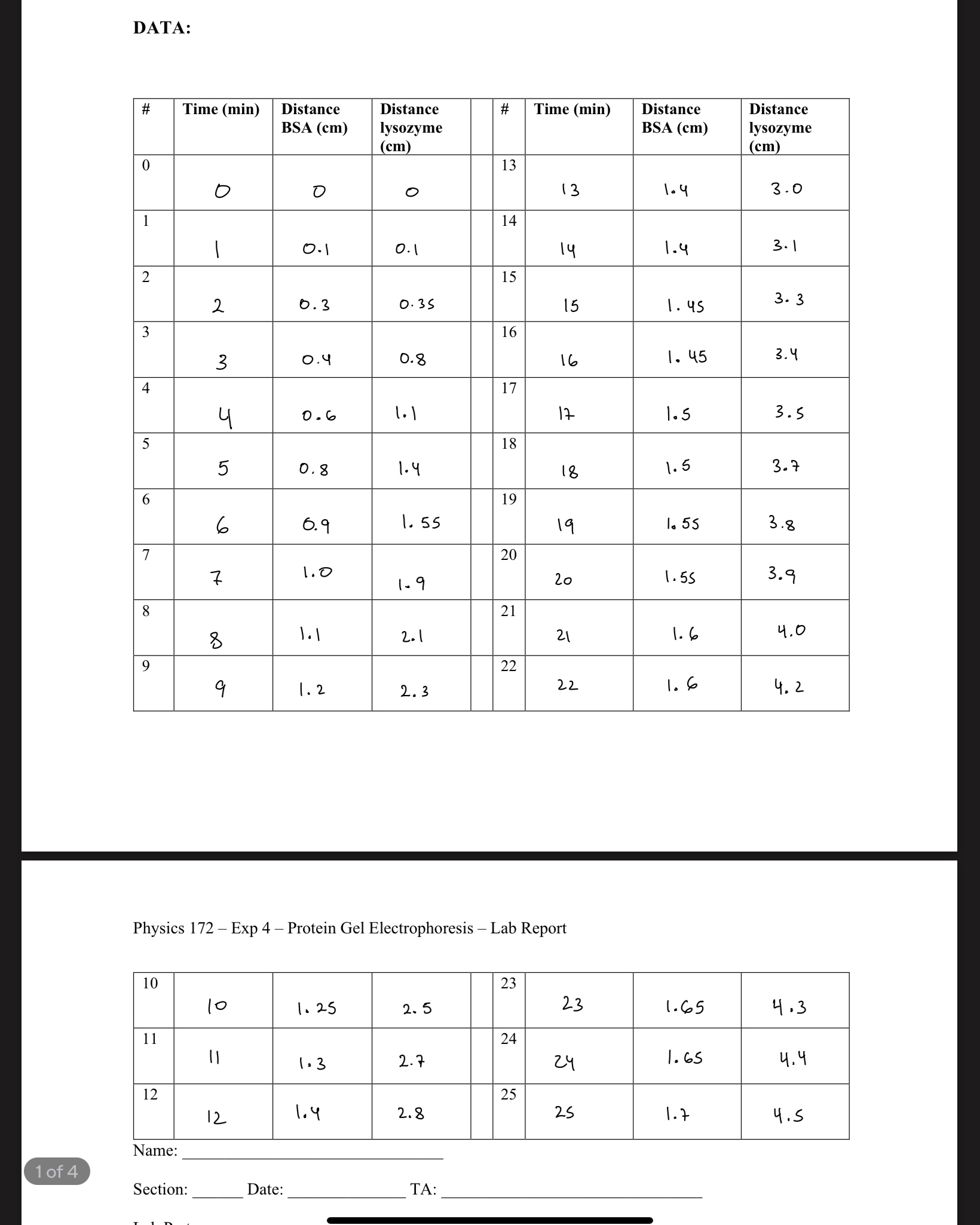
Question: CALCULATIONS 2. +/- R2 = Vterm,BSA +/- R2 = Vterm, lysozyme Vterm,BSA 3. = Vterm,lysozyme M = M 4. % diff = [ratioexp - ratioacc

Step by Step Solution
There are 3 Steps involved in it
1 Expert Approved Answer
Step: 1 Unlock


Question Has Been Solved by an Expert!
Get step-by-step solutions from verified subject matter experts
Step: 2 Unlock
Step: 3 Unlock


