Question: Data handling Interpret the results presented in the provided figures to answer the questions below. In the Neurogenetics Research Facility, the team has been investigating
Data handling
Interpret the results presented in the provided figures to answer the questions below.
In the Neurogenetics Research Facility, the team has been investigating a unique mouse model exhibiting progressive neurological decline reminiscent of human neurodegenerative diseases.
A mutated gene, named lpg surfaced as a candidate of interest due to its sequence similarity to deubiquitinating enzymes DUBs
To explore Ipg function, the team has conducted an analysis of the corresponding enzyme Ipg while particularly focusing on its interaction with PCNA, a protein known to be ubiquitinated in response to cellular stress. The goal is to discern the role of
a Review Figure which depicts a ubiquitination assay involving PCNA and a mutant variant. Outline the method used to monitor ubiquitination and infer the significance of observed molecular weight changes in PCNA over the indicated time intervals.
b Examine Figure showcasing a deubiquitination assay poststress induction.
Discuss how the experiment tracks changes in PCNA and what the temporal alterations suggest about the deubiquitination mechanism in response to DNA damage.
c Consider Figure detailing an enzymatic activity assay.
Describe the process used to assess the enzyme's specificity for different ubiquitin chains.
Analyse the cleavage patterns to determine the enzyme's activity profile.
d Reflect on the potential link between the enzymatic function of lpg and the neurological symptoms observed in the mouse model.
Contemplate the broader implications of ubiquitination dynamics in neuronal health.
Abstract:
Based on the provided Figures and your analysis, construct an abstract to summarise the study. The abstract needs to include a title, background, methods, results and conclusion.
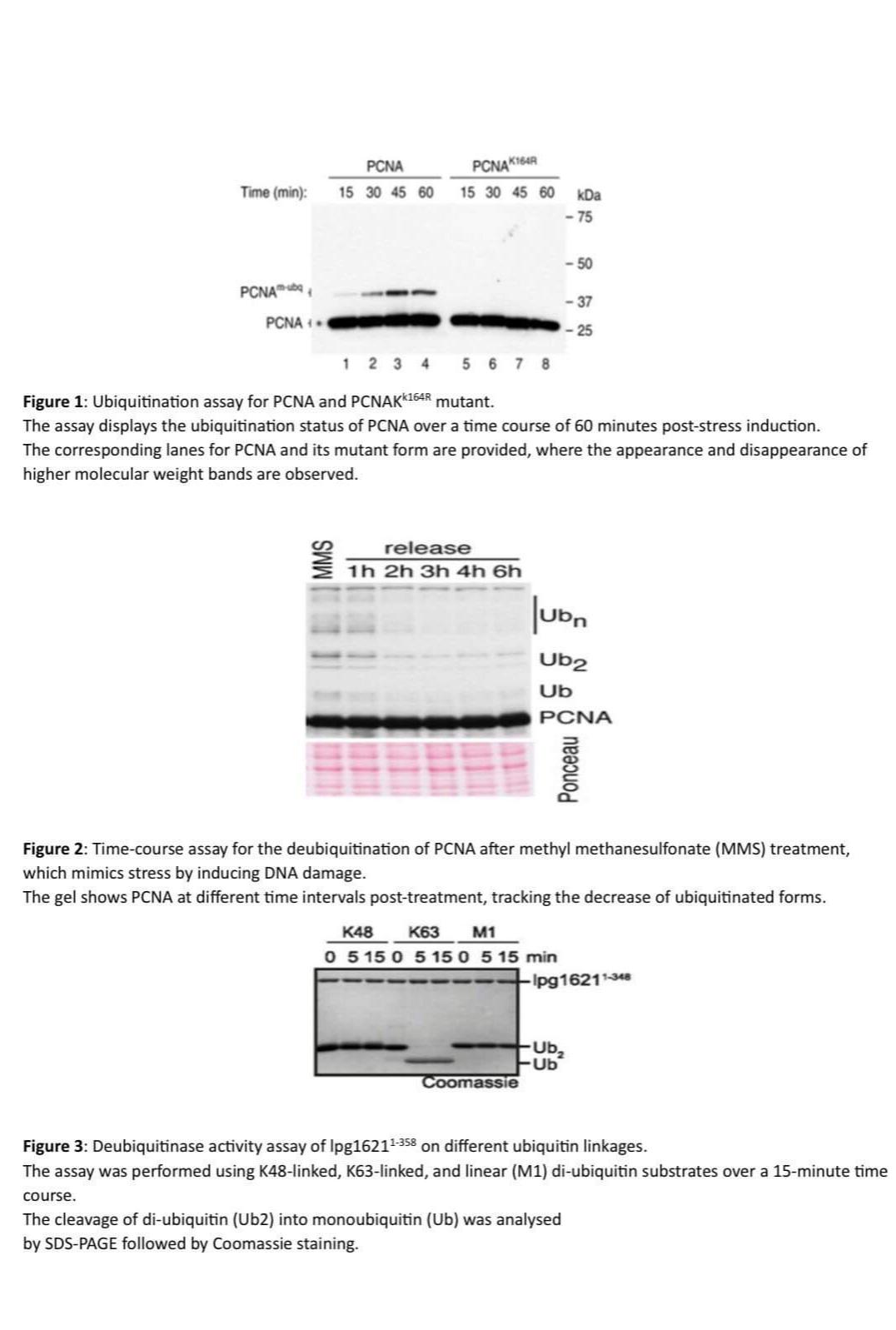
Figure : Ubiquitination assay for PCNA and PCNAK mutant.
The assay displays the ubiquitination status of PCNA over a time course of minutes poststress induction. The corresponding lanes for PCNA and its mutant form are provided, where the appearance and disappearance of higher molecular weight bands are observed.
Figure : Timecourse assay for the deubiquitination of PCNA after methyl methanesulfonate MMS treatment, which mimics stress by inducing DNA damage.
The gel shows PCNA at different time intervals posttreatment, tracking the decrease of ubiquitinated forms.
Figure : Deubiquitinase activity assay of lpg on different ubiquitin linkages.
The assay was performed using Klinked, Klinked, and linear M diubiquitin substrates over a minute time course.
The cleavage of diubiquitin Ub into monoubiquitin Ub was analysed by SDSPAGE followed by Coomassie staining.
Step by Step Solution
There are 3 Steps involved in it
1 Expert Approved Answer
Step: 1 Unlock


Question Has Been Solved by an Expert!
Get step-by-step solutions from verified subject matter experts
Step: 2 Unlock
Step: 3 Unlock


