Question: fQuestion 4: (2 marks) Did the gene with the largest difference of means (in terms of mag- nitude/absolute value) get the smallest p-value? Explain why
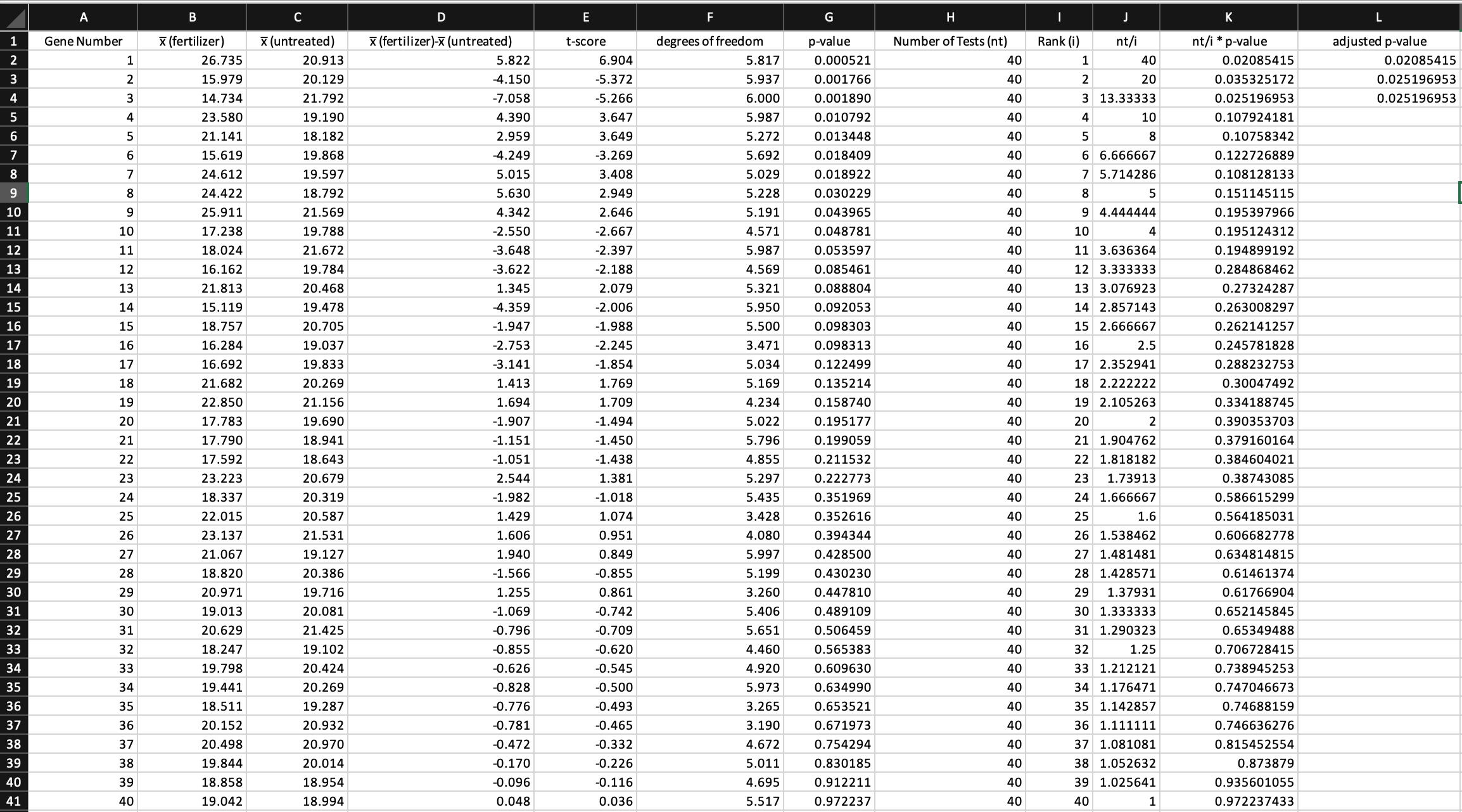

\fQuestion 4: (2 marks) Did the gene with the largest difference of means (in terms of mag- nitude/absolute value) get the smallest p-value? Explain why this might not be an error, and which piece(s) of missing information could settle the issue. (Hint: How is the t-score calculated?) Question 5: (1 mark) Using an 0: threshold of 0.05, which genes are showing statistically signicant differences? Question 6: (2 marks) If none of these genes were in truth aected by the treatment (i.e. if the null hypothesis of no difference were true for all genes), how many genes would you expect to show statistically signicant differences, on average, if a threshold of o: = 0.05 were used? Justify your answer by describing the number of statistically signicant p-values using a Binomial distribution and determining its expected value. Question 7: (2 marks) If none of these genes were in truth affected by the treatment (i.e. if the null hypothesis of no difference were true for all genes) and a threshold of a = 0.05 were used, what would the family-wise error rate be for this set of genes? Justify your answer by describing the number of statistically signicant pvalues using a Binomial distribution. Question 8: (2 marks) Use the Bonferroni correction to determine an alternative pvalue threshold that will control the family-wise error rate at 0.05. Do any genes show statistically signicant differences using this threshold? Question 9: (3 marks) Use the BenjaminiHochberg method to determine False-Discovery- rateadjusted p-values for the genes in this data set. The provided spreadsheet has provided some preliminary work towards this goal, and has correct adjusted pvalues for the rst three genes. Provide the completed list of 40 adjusted pvalues, in their current order, as part of your report. Question 10: (2 marks) Suppose your sponsor is willing to fund a follow-up experiment to validate your ndings, but wants you to make sure that no more than a quarter of genes you nominate for validation are false positives (on average). Explain how you could determine this using your adjusted pvalues, and provide the \"Gene Numbers\" for the list of candidates to validate
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


