Question: Here is my python code that translates DNA to protein... I need to now find out how many 'A' 'L' and 'H' are in the
Here is my python code that translates DNA to protein... I need to now find out how many 'A' 'L' and 'H' are in the protein sequence... What do I need to change or add to this code to do that?
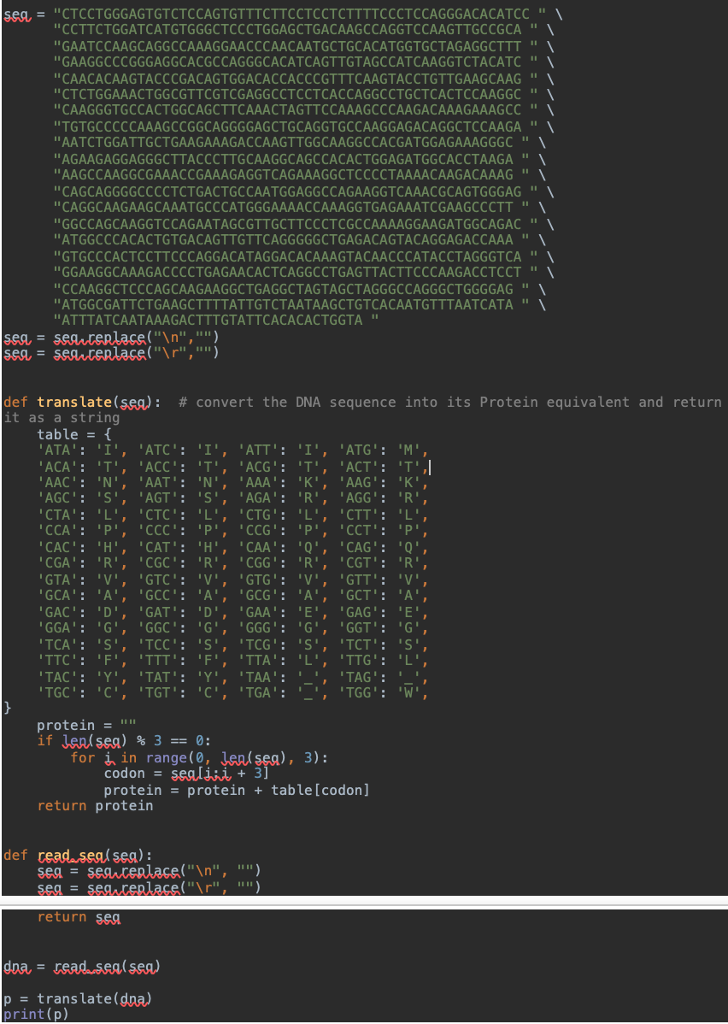
seq = "CTCCTGGGAGTGTCTCCAGTGTTTCTTCCTCCTCTTTTCCCTCCAGGGACACATCC " \ "CCTTCTGGATCATGTGGGCTCCCTGGAGCTGACAAGCCAGGTCCAAGTTGCCGCA " \ "GAATCCAAGCAGGCCAAAGGAACCCAACAATGCTGCACATGGTGCTAGAGGCTTT " \ "GAAGGCCCGGGAGGCACGCCAGGGCACATCAGTTGTAGCCATCAAGGTCTACATC " \ "CAACACAAGTACCCGACAGTGGACACCACCCGTTTCAAGTACCTGTTGAAGCAAG " \ "CTCTGGAAACTGGCGTTCGTCGAGGCCTCCTCACCAGGCCTGCTCACTCCAAGGC " \ "CAAGGGTGCCACTGGCAGCTTCAAACTAGTTCCAAAGCCCAAGACAAAGAAAGCC " \ "TGTGCCCCCAAAGCCGGCAGGGGAGCTGCAGGTGCCAAGGAGACAGGCTCCAAGA " \ "AATCTGGATTGCTGAAGAAAGACCAAGTTGGCAAGGCCACGATGGAGAAAGGGC " \ "AGAAGAGGAGGGCTTACCCTTGCAAGGCAGCCACACTGGAGATGGCACCTAAGA " \ "AAGCCAAGGCGAAACCGAAAGAGGTCAGAAAGGCTCCCCTAAAACAAGACAAAG " \ "CAGCAGGGGCCCCTCTGACTGCCAATGGAGGCCAGAAGGTCAAACGCAGTGGGAG " \ "CAGGCAAGAAGCAAATGCCCATGGGAAAACCAAAGGTGAGAAATCGAAGCCCTT " \ "GGCCAGCAAGGTCCAGAATAGCGTTGCTTCCCTCGCCAAAAGGAAGATGGCAGAC " \ "ATGGCCCACACTGTGACAGTTGTTCAGGGGGCTGAGACAGTACAGGAGACCAAA " \ "GTGCCCACTCCTTCCCAGGACATAGGACACAAAGTACAACCCATACCTAGGGTCA " \ "GGAAGGCAAAGACCCCTGAGAACACTCAGGCCTGAGTTACTTCCCAAGACCTCCT " \ "CCAAGGCTCCCAGCAAGAAGGCTGAGGCTAGTAGCTAGGGCCAGGGCTGGGGAG " \ "ATGGCGATTCTGAAGCTTTTATTGTCTAATAAGCTGTCACAATGTTTAATCATA " \ "ATTTATCAATAAAGACTTTGTATTCACACACTGGTA "
seq = seq.replace(" ","")
seq = seq.replace(" ","")
def translate(seq): # convert the DNA sequence into its Protein equivalent and return it as a string
table = { 'ATA': 'I', 'ATC': 'I', 'ATT': 'I', 'ATG': 'M', 'ACA': 'T', 'ACC': 'T', 'ACG': 'T', 'ACT': 'T', 'AAC': 'N', 'AAT': 'N', 'AAA': 'K', 'AAG': 'K', 'AGC': 'S', 'AGT': 'S', 'AGA': 'R', 'AGG': 'R', 'CTA': 'L', 'CTC': 'L', 'CTG': 'L', 'CTT': 'L', 'CCA': 'P', 'CCC': 'P', 'CCG': 'P', 'CCT': 'P', 'CAC': 'H', 'CAT': 'H', 'CAA': 'Q', 'CAG': 'Q', 'CGA': 'R', 'CGC': 'R', 'CGG': 'R', 'CGT': 'R', 'GTA': 'V', 'GTC': 'V', 'GTG': 'V', 'GTT': 'V', 'GCA': 'A', 'GCC': 'A', 'GCG': 'A', 'GCT': 'A', 'GAC': 'D', 'GAT': 'D', 'GAA': 'E', 'GAG': 'E', 'GGA': 'G', 'GGC': 'G', 'GGG': 'G', 'GGT': 'G', 'TCA': 'S', 'TCC': 'S', 'TCG': 'S', 'TCT': 'S', 'TTC': 'F', 'TTT': 'F', 'TTA': 'L', 'TTG': 'L', 'TAC': 'Y', 'TAT': 'Y', 'TAA': '_', 'TAG': '_', 'TGC': 'C', 'TGT': 'C', 'TGA': '_', 'TGG': 'W', }
protein = ""
if len(seq) % 3 == 0:
for i in range(0, len(seq), 3):
codon = seq[i:i + 3]
protein = protein + table[codon]
return protein
def read_seq(seq):
seq = seq.replace(" ", "")
seq = seq.replace(" ", "")
return seq
dna = read_seq(seq)
p = translate(dna)
print(p)
sea-"CTCCTGGGAGTGTCTCCAGTGTTTCTTCCTCCTCTTTTCCCTCCAGGGACACATCC " "CCTTCTGGATCATGTGGGCTCCCTGGAGCTGACAAGCCAGGTCCAAGTTGCCGCA "GAATCCAAGCAGGCCAAAGGAACCCAACAATGCTGCACATGGTGCTAGAGGCTTT" "GAAGGCCCGGGAGGCACGCCAGGGCACATCAGTTGTAGCCATCAAGGTCTACATC "CAACACAAGTACCCGACAGTGGACACCACCCGTTTCAAGTACCTGTTGAAGCAAG "CTCTGGAAACTGGCGTTCGTCGAGGCCTCCTCACCAGGCCTGCTCACTCCAAGGC "CAAGGGTGCCACTGGCAGCTTCAAACTAGTTCCAAAGCCCAAGACAAAGAAAGCC "TGTGCCCCCAAAGCCGGCAGGGGAGCTGCAGGTGCCAAGGAGACAGGCTCCAAGA AATCTGGATTGCTGAAGAAAGACCAAGTTGGCAAGGCCACGATGGAGAAAGGGC" "AGAAGAGGAGGGCTTACCCTTGCAAGGCAGCCACACTGGAGATGGCACCTAAGA "AAGCCAAGGCGAAACCGAAAGAGGTCAGAAAGGCTCCCCTAAAACAAGACAAAG "CAGCAGGGGCCCCTCTGACTGCCAATGGAGGCCAGAAGGTCAAACGCAGTGGGAG "CAGGCAAGAAGCAAATGCCCATGGGAAAACCAAAGGTGAGAAATCGAAGCCCTT " "GGCCAGCAAGGTCCAGAATAGCGTTGCTTCCCTCGCCAAAAGGAAGATGGCAGAC "ATGGCCCACACTGTGACAGTTGTTCAGGGGGCTGAGACAGTACAGGAGACCAAA "GTGCCCACTCCTTCCCAGGACATAGGACACAAAGTACAACCCATACCTAGGGTCA "GGAAGGCAAAGACCCCTGAGAACACTCAGGCCTGAGTTACTTCCCAAGACCTCCT" "CCAAGGCTCCCAGCAAGAAGGCTGAGGCTAGTAGCTAGGGCCAGGGCTGGGGAG ATGGCGATTCTGAAGCTTTTATTGTCTAATAAGCTGTCACAATGTTTAATCATA "ATTTATCAATAAAGACTTTGTATTCACACACTGGTA def translate(sea): # convert the DNA sequence into its Protein equivalent and return it as a string CTA': L CTC': 'L',CTG': 'L,"CTT':'L' GCA': 'A,'GCC':'A,'GCG': 'A, 'GCT': A' GAC': 'D'GAT':'D 'GAA':EGAG':'E', if protein protein table [codon] return protein sea seasea,replace("r",)
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


