Question: I created a data matrix; I need help expressing the relationships among taxa marking the derived character states at the node where they change. .
I created a data matrix; I need help expressing the relationships among taxa marking the derived character states at the node where they change. . # 1 # 2 #3 #4 #5 #6 #7 #8 #9 1. #10 #11.
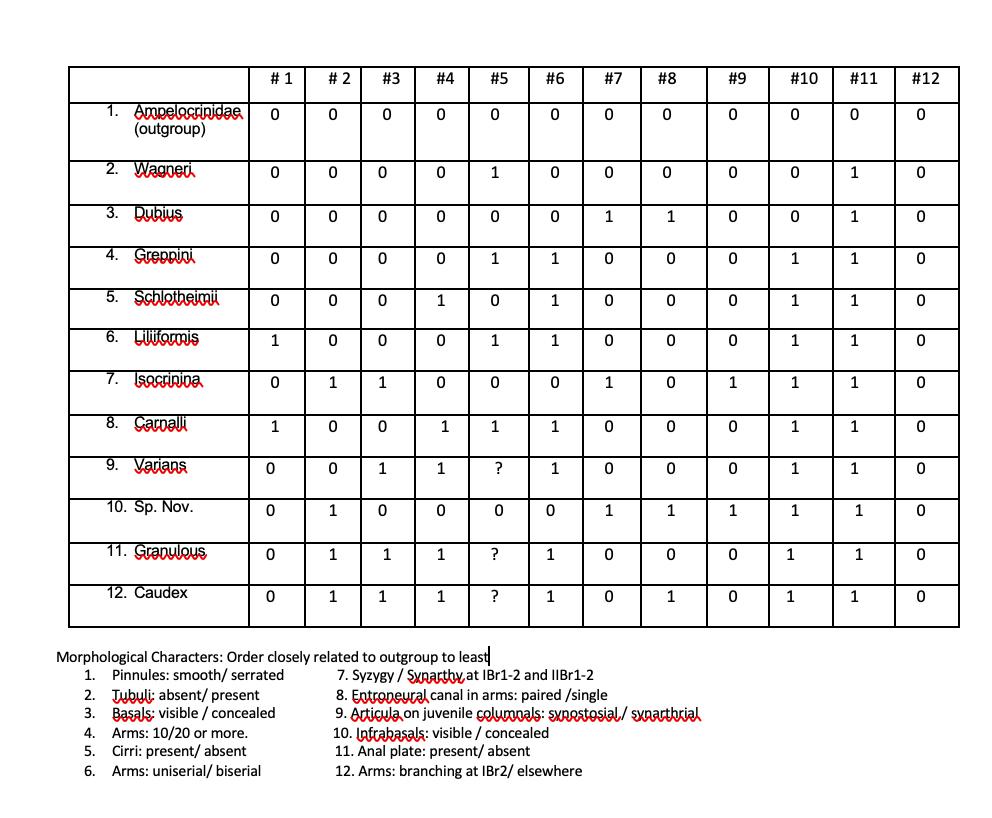
1. Ampelacrinidae (outgroup) 2. Wagneri 3. Dubius 4. Greppini 5. Schlotheimi 6. Litfarmis 7. Isacrinina 8. Carballi 9. Varians 10. Sp. Nov. 11. Granulous 12. Caudex # 1 0 0 Ol 0 0 1 0 1 0 0 0 0 # 2 0 ol 0 lo 0 0 0 0 1 0 0 1 1 1 #3 0 0 0 0 0 0 1 0 1 ol 1 1 #4 0 0 P1 0 0 1 ol 0 1 1 0 1 1 #5 0 1 0 1 0 1 0 1 ? Morphological Characters: Order closely related to outgroup to least 1. Pinnules: smooth/ serrated 2. Tubuli: absent/present 3. Baals: visible / concealed 4. Arms: 10/20 or more. 5. Cirri: present/absent 6. Arms: uniserial/ biserial 0 ? ? #6 0 0 lo 0 1 1 1 0 1 1 0 1 1 7. Syzygy/Sxnarthy at IBr1-2 and IIBr1-2 #7 10. Infrabasals: visible / concealed 11. Anal plate: present/absent 12. Arms: branching at IBr2/ elsewhere 0 lo 0 1 0 To 0 lo 0 1 0 0 1 lo 0 lo 0 #8 lo 0 0 1 0 la 0 lo 0 ol 0 lo 0 lo 0 1 0 1 8. Entroneural canal in arms: paired /single 9. Articula on juvenile columnals: sxnostosial/sxnarthrial #9 0 0 0 0 0 0 1 0 0 1 0 0 # 10 0 0 lo 0 1 1 1 1 1 1 1 1 #11 0 1 L 1 1 1 1 1 1 1 1 1 1 #12 0 0 0 0 0 lo 0 lo 0 0 0 la 0 lo 0 la 0
Step by Step Solution
3.43 Rating (156 Votes )
There are 3 Steps involved in it
ANSWER To express the relationships among taxa marking the derived character states at the node where they change we can analyze the provided data mat... View full answer

Get step-by-step solutions from verified subject matter experts


