Question: in python please please show all steps and the output too thanks The starter code can be found in the file **(get_fused_sequence_conplement.pyl(get_fused_sequence_complement.py)**. Given two DNA
in python please
please show all steps and the output too
thanks
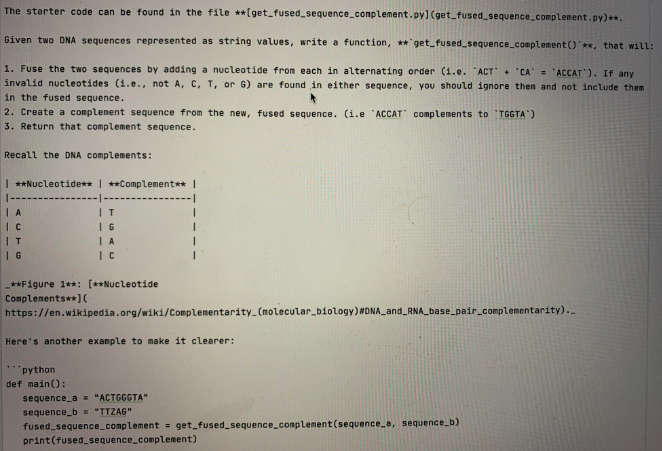
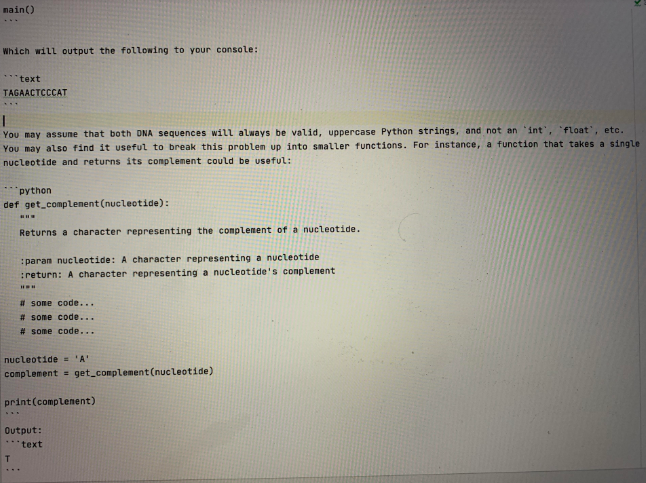
The starter code can be found in the file **(get_fused_sequence_conplement.pyl(get_fused_sequence_complement.py)**. Given two DNA sequences represented as string values, write a function, ** get_fused_sequence_complement('**, that will: 1. Fuse the two sequences by adding a nucleotide from each in alternating order (1.e. "ACT' + 'CA' - 'ACCAT"). If any invalid nucleotides (i.e., not A, C, T, or G) are found in either sequence, you should ignore then and not include them in the fused sequence. 2. Create a complement sequence from the new, fused sequence. (i.e 'ACCAT' complements to TGGTA) 3. Return that complement sequence. Recall the DNA complements: | **Nucleotide** | **Complement** | IT IG IA IC IT IG **Figure 1**: [**Nucleotide Complements**] https://en.wikipedia.org/wiki/Complementarity_(molecular biology) MONA_and_RNA_base_pair_complementarity).. Here's another example to make it clearer: python def main(): sequence_a - "ACTGEGTA" sequence_b - "TTZAG" fused_sequence_complement = get_fused_sequence complement(sequence_., sequence_b) print(fused_sequence_complement) nain() Which will output the following to your console: "text TAGAACTCCCAT You may assune that both DNA sequences will always be valid, uppercase Python strings, and not an 'int', 'float', etc. You may also find it useful to break this problem up into smaller functions. For instance, a function that takes a single nucleotide and returns its complement could be useful: python def get_complement(nucleotide): Returns a character representing the complement of a nucleotide. paran nucleotide: A character representing a nucleotide return: A character representing a nucleotide's complenent HOW # some code... # some code... # some code... nucleotide = 'A' complement = get_complement(nucleotide) print(complement) Output: text T
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


