Question: LAB TASK: Implement a program in python that will open a FASTA file, concatenate its multiline sequences into single strings, store them in a dictionary
LAB TASK: Implement a program in python that will open a FASTA file, concatenate its multiline sequences into single strings, store them in a dictionary using the sequence ID from the sequence header (value between the | symbols) as a key, and then print the IDs and sequences as two columns in a new file.
OBJECTIVE(S):
1. Write your code in the block below. Download the file called myoglobin.fasta, and make sure to save it in the same location as your lab task script.
2. Create an empty dictionary to store sequence information.
3. Using the open function, open the FASTA file (myoglobin.fasta).
4. When you find a line beginning with the > character (a header) extract the ID code between the | symbols and start a new dictionary entry using the ID as a key.
5. If a line isnt a header (i.e. it is a sequence), strip off the newline character at the end and append the sequence to a growing string (to the growing sequence that is the dictionary value) stored within the most recent dictionary key.
6. Close the original file.
7. Open a new file for writing, e.g. myoglobin_processed.txt.
8. Loop through the dictionary and write the ID keys and their corresponding sequences to the new file, separating them with a tab (\t) to generate two columns.
9. Close the new file.
10. Run your script. Upload the script and output (myoglobin_processed.txt) for lab credit. Dont forget comments!
Expected output for two sequences should look like this (note how the sequence now is a single string):
P02189 MGLSDGEWQLVLNVWGKVEADVAGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASEDLKKHGNTVLTALGGILKKKGHHEAELTPLAQSHATKHKIPVKYLEFISEAIIQVLQSKHPGDFGADAQGAMSKALELFRNDMAAKYKELGFQG P04247 MGLSDGEWQLVLNVWGKVEADLAGHGQEVLIGLFKTHPETLDKFDKFKNLKSEEDMKGSEDLKKHGCTVLTALGTILKKKGQHAAEIQPLAQSHATKHKIPVKYLEFISEIIIEVLKKRHSGDFGADAQGAMSKALELFRNDIAAKYKELGFQG
The provided myoglobin.fasta file I have to work with contains the following:
>sp|P02192|MYG_BOVIN Myoglobin OS=Bos taurus GN=MB PE=1 SV=3 MGLSDGEWQLVLNAWGKVEADVAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASE DLKKHGNTVLTALGGILKKKGHHEAEVKHLAESHANKHKIPVKYLEFISDAIIHVLHAKH PSDFGADAQAAMSKALELFRNDMAAQYKVLGFHG >sp|P02189|MYG_PIG Myoglobin OS=Sus scrofa GN=MB PE=1 SV=2 MGLSDGEWQLVLNVWGKVEADVAGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASE DLKKHGNTVLTALGGILKKKGHHEAELTPLAQSHATKHKIPVKYLEFISEAIIQVLQSKH PGDFGADAQGAMSKALELFRNDMAAKYKELGFQG >sp|P02144|MYG_HUMAN Myoglobin OS=Homo sapiens GN=MB PE=1 SV=2 MGLSDGEWQLVLNVWGKVEADIPGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASE DLKKHGATVLTALGGILKKKGHHEAEIKPLAQSHATKHKIPVKYLEFISECIIQVLQSKH PGDFGADAQGAMNKALELFRKDMASNYKELGFQG >sp|P68082|MYG_HORSE Myoglobin OS=Equus caballus GN=MB PE=1 SV=2 MGLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASE DLKKHGTVVLTALGGILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISDAIIHVLHSKH PGDFGADAQGAMTKALELFRNDIAAKYKELGFQG >sp|P04247|MYG_MOUSE Myoglobin OS=Mus musculus GN=Mb PE=1 SV=3 MGLSDGEWQLVLNVWGKVEADLAGHGQEVLIGLFKTHPETLDKFDKFKNLKSEEDMKGSE DLKKHGCTVLTALGTILKKKGQHAAEIQPLAQSHATKHKIPVKYLEFISEIIIEVLKKRH SGDFGADAQGAMSKALELFRNDIAAKYKELGFQG >sp|P02197|MYG_CHICK Myoglobin OS=Gallus gallus GN=MB PE=1 SV=4 MGLSDQEWQQVLTIWGKVEADIAGHGHEVLMRLFHDHPETLDRFDKFKGLKTPDQMKGSE DLKKHGATVLTQLGKILKQKGNHESELKPLAQTHATKHKIPVKYLEFISEVIIKVIAEKH AADFGADSQAAMKKALELFRNDMASKYKEFGFQG
This is my code so far:
file = open('myoglobin.fasta','r') l={} for line in file: if line[0]=='>': m=line.split("|") m[2]=m[2].rstrip() l[m[1]]=m[2] c=m[1] else: line=line.rstrip() l[c]=l[c]+line file.close() out=open('myoglobin_processed.txt','w') for key in l: out.write(key) out.write("\t") out.write(l[key]) out.write(" ") out.close()
This is my current output, but it's not correct. I don't know how to get it to look like the expected output which is described above. Any help would be great!
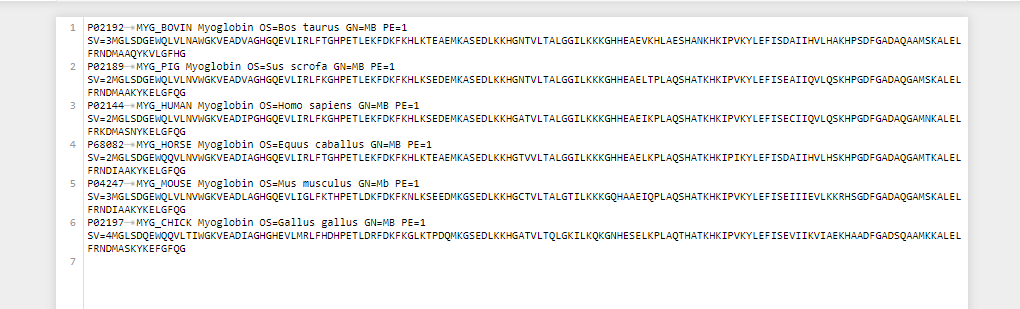
1 P02192- MYG_BOVIN Myoglobin OS=Bos taurus GN=MB PE=1 SV=3MGLSDGEWOLVL NAWGKVEADVAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASEDLKKHGNTVLTALGGILKKKGHHEAEVKHLAESHANKHKIPVKYLEFISDAITHVLHAKHPSDFGADAQAAMSKALEL FRNDMAAQYKVLGFHG 2 P02189 MYG_PIG Myoglobin Os=Sus scrofa GN=MB PE=1 SV=2MGLSDGEWQLVLNVWGKVEADVAGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASEDLKKHGNTVLTALGGILKKKGHHEAELTPLAQSHATKHKIPVKYLEFISEAIIQVLOSKHPGDFGADAQGAMSKALEL FRNDMAAKYKELGFOG P02144 MYG_HUMAN Myoglobin OS=Homo sapiens GN=MB PE=1 SV=2MGL SDGEWQLVLNVWGKVEADIPGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASEDLKKHGATVLTALGGILKKKGHHEAEIKPLAQSHATKHKIPVKYLEFISECIIQVLOSKHPGDFGADAQGAMNKALEL FRKDMASNYKELGFOG P68082- MYG_HORSE Myoglobin os=Equus caballus GN=MB PE-1 SV=2MGLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASEDLKKHGTVLTALGGILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISDAITHVLHSKHPGDFGADAQGAMTKALEL FRNDIAAKYKELGFOG 5 P04247 MYG_MOUSE Myoglobin OS-Mus musculus GN=Mb PE=1 SV=3MGLSDGEWQLVL NVWGKVEADLAGHGQEVLIGLFKTHPETLDKFDKFKNLKSEEDMKGSEDLKKHGCTVLTALGTILKKKGQHAAE IOPLAQSHATKHKIPVKYLEFISEIIIEVLKKRHSGDFGADAQGAMSKALEL FRNDIAAKYKELGFOG 6 P02197- MYG_CHICK Myoglobin OS-Gallus gallus GN=MB PE=1 SV=4MGLSDQEWQQVLTIWGKVEADIAGHGHEVLMRLFHDHPETLDRFDKFKGLKTPDOMKGSEDLKKHGATVLTQLGKILKQKGNHESELKPLAQTHATKHKIPVKYLEFISEVIIKVIAEKHAADFGADSQAAMKKALEL FRNDMASKYKEFGFQG
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


