Question: Learning objectives Write a function that produces two outputs. .Use strcat to concatenate strings. Use array indexing and the colon operator (:) to extract parts
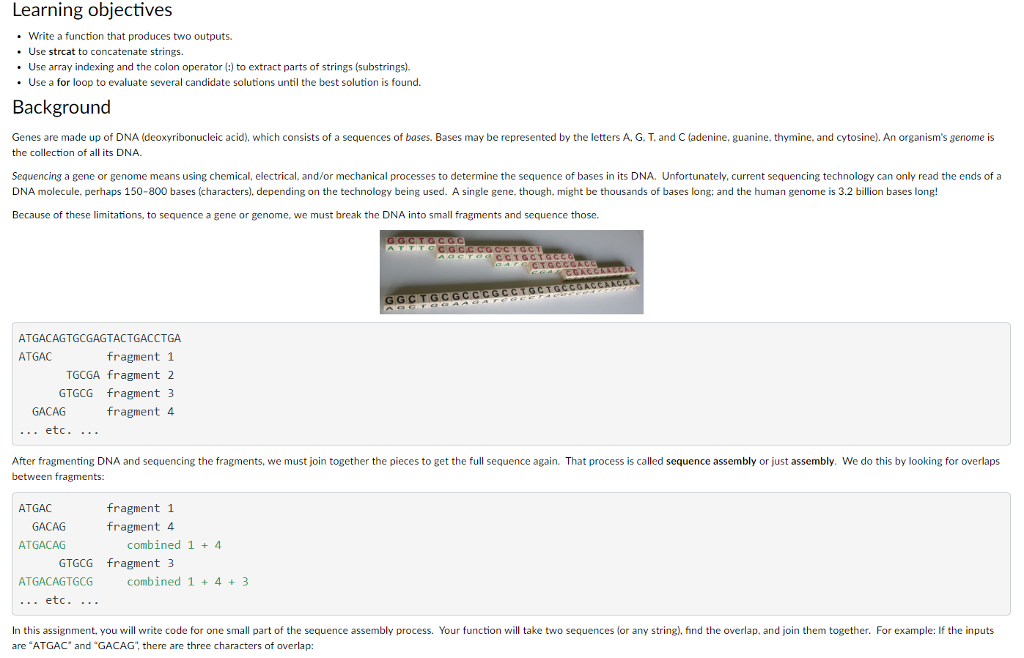
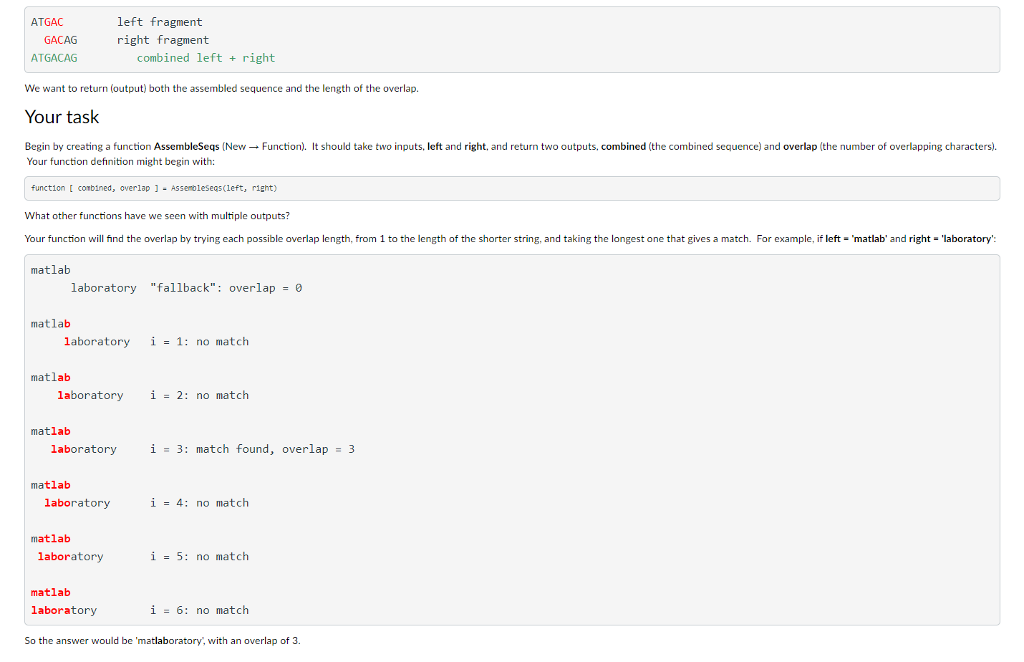
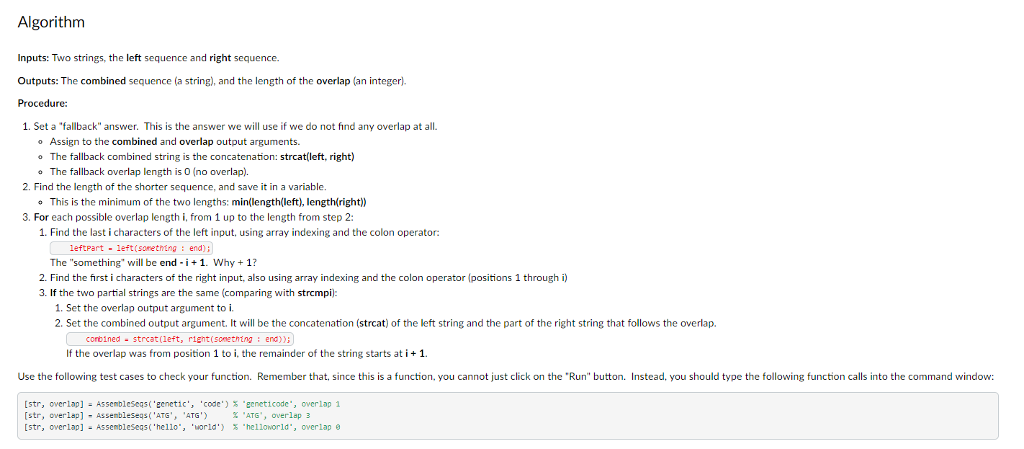
Learning objectives Write a function that produces two outputs. .Use strcat to concatenate strings. Use array indexing and the colon operator (:) to extract parts of strings (substrings). . Use a for loop to evaluate several candidate solutions until the best solution is found. Background Genes are made up of DNA (deoxyribonucleic acid), which consists of a sequences of bases. Bases may be represented by the letters A. G. T, and C (adenine, guanine, thymine, and cytosine). An organism's genome is the collection of all its DNA. Sequencing a gene or genome means using chemical, electrical, and/or mechanical processes to determine the sequence of bases in its DNA. Unfortunately, current sequencing technology can only read the ends of a DNA molecule, perhaps 150-800 bases (characters), depending on the technology being used. A single gene, though, might be thousands of bases long: and the human genome is 3.2 billion bases long! Because of these limitations, to sequence a gene or genome, we must break the DNA into small fragments and sequence those. GCTGGC ATGACAGTGCGAGTACTGACCTGA ATGAC fragment 1 TGCGA fragment 2 GTGCG fragment3 fragment 4 GACAG etc. .. . After fragmenting DNA and sequencing the fragments, we must join together the pieces to get the full sequence again. That process is called sequence assembly or just assembly. We do this by looking for overlaps between fragments: fragment 1 fragment 4 ATGAC GACAG ATGACAG combined 1 4 GTGCG fragment3 ATGACAGTGCG combined 1 + 43 etc. .. . In this assignment, you will write code for one small part of the sequence assembly process. Your function will take two sequences (or any string), find the overlap, and join them together. For example: If the inputs are "ATGAC and "GACAG, there are three characters of overlap: Learning objectives Write a function that produces two outputs. .Use strcat to concatenate strings. Use array indexing and the colon operator (:) to extract parts of strings (substrings). . Use a for loop to evaluate several candidate solutions until the best solution is found. Background Genes are made up of DNA (deoxyribonucleic acid), which consists of a sequences of bases. Bases may be represented by the letters A. G. T, and C (adenine, guanine, thymine, and cytosine). An organism's genome is the collection of all its DNA. Sequencing a gene or genome means using chemical, electrical, and/or mechanical processes to determine the sequence of bases in its DNA. Unfortunately, current sequencing technology can only read the ends of a DNA molecule, perhaps 150-800 bases (characters), depending on the technology being used. A single gene, though, might be thousands of bases long: and the human genome is 3.2 billion bases long! Because of these limitations, to sequence a gene or genome, we must break the DNA into small fragments and sequence those. GCTGGC ATGACAGTGCGAGTACTGACCTGA ATGAC fragment 1 TGCGA fragment 2 GTGCG fragment3 fragment 4 GACAG etc. .. . After fragmenting DNA and sequencing the fragments, we must join together the pieces to get the full sequence again. That process is called sequence assembly or just assembly. We do this by looking for overlaps between fragments: fragment 1 fragment 4 ATGAC GACAG ATGACAG combined 1 4 GTGCG fragment3 ATGACAGTGCG combined 1 + 43 etc. .. . In this assignment, you will write code for one small part of the sequence assembly process. Your function will take two sequences (or any string), find the overlap, and join them together. For example: If the inputs are "ATGAC and "GACAG, there are three characters of overlap
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


