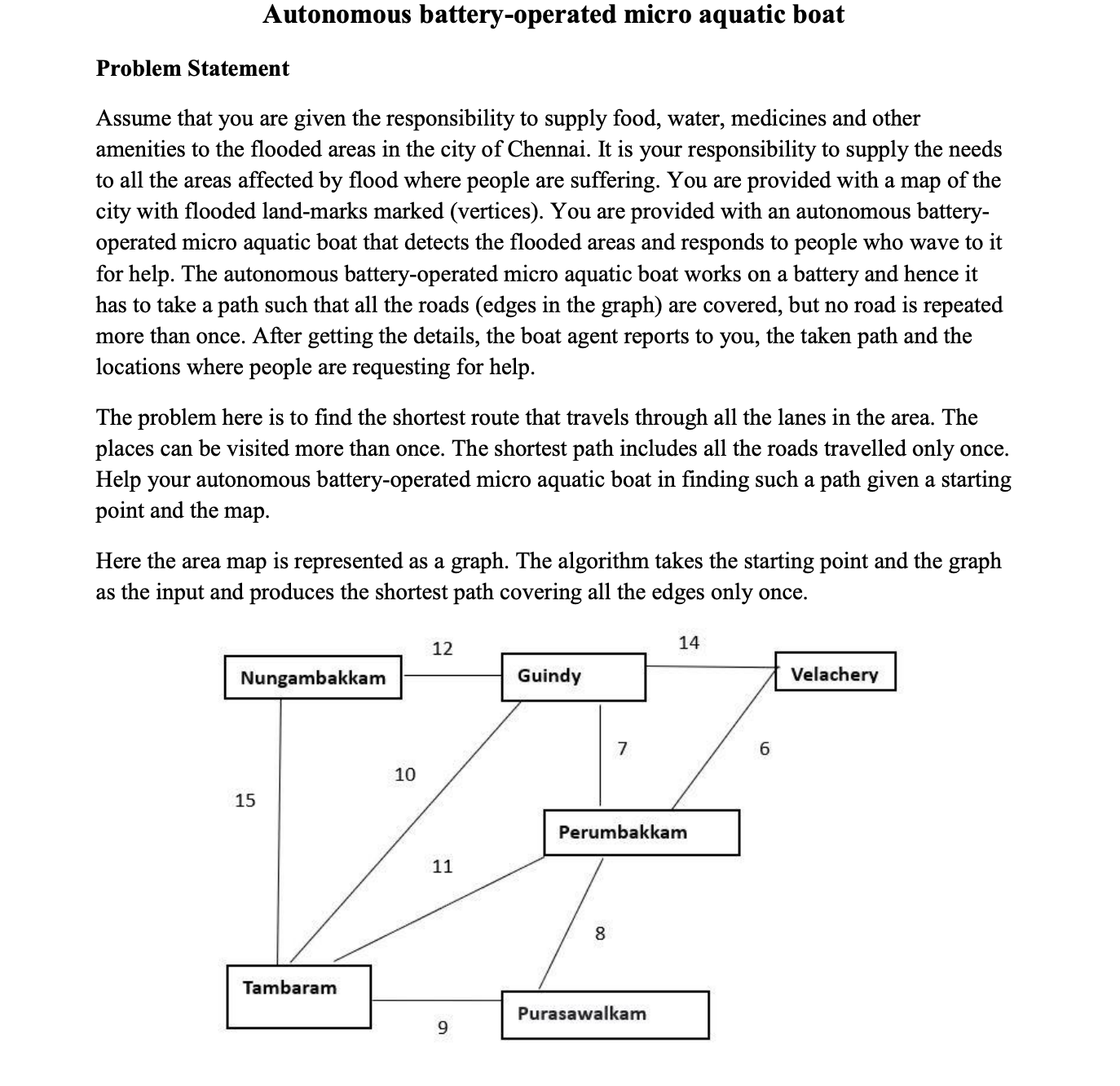
Question: Modify the program below, to get the best path which are connected as per diagram. Currently program below gives me best path where one node
Modify the program below, to get the best path which are connected as per diagram. Currently program below gives me best path where one node to next as no straight connection. For example, one such Best Path my program giving is: pupetvgn where t tambaram has no straight connection with vvelachery Please correct my genetic algorithm given
Genetic Algorithm for the attached problem is as follow It will run without error
import random
# Node coordinates as provided
nodecoordinates
pu:
pe:
t:
v:
g:
n:
# Function to calculate Manhattan distance between two points
def manhattandistancepoint point:
x y nodecoordinatespoint
x y nodecoordinatespoint
return absx x absy y
# Function to generate the initial population of paths
def generateinitialpopulationgraph populationsize:
initialpopulation
for in rangepopulationsize:
path listgraphkeys
random.shufflepath
initialpopulation.appendpath
return initialpopulation
# Function to calculate fitness of a path
def calculatefitnesspath:
totaldistance summanhattandistancepathi pathi for i in rangelenpath
return totaldistance if totaldistance else floatinf
# Genetic Algorithm components
def roulettewheelselectionpopulation fitnesses:
totalfitness sumfitnesses
pick random.uniform totalfitness
current
for individual, fitness in zippopulation fitnesses:
current fitness
if current pick:
return individual
def crossoverparent parent split'single':
if split 'single':
point random.randint lenparent
child parent:pointgene for gene in parent if gene not in parent:point
child parent:pointgene for gene in parent if gene not in parent:point
else: # Twopoint crossover
point point sortedrandomsamplerange lenparent
child parent:pointgene for gene in parentpoint:point if gene not in parent:point parentpoint:
child parent:pointgene for gene in parentpoint:point if gene not in parent:point parentpoint:
return child child
def mutatepath mutationrate:
for i in rangelenpath:
if random.random mutationrate:
swapwith random.randint lenpath
pathi pathswapwith pathswapwith pathi
return path
# Main Genetic Algorithm function
def geneticalgorithmgraph populationsize, generations, mutationrate:
population generateinitialpopulationgraph populationsize
for in rangegenerations:
fitnesses calculatefitnessindividual for individual in population
newpopulation
while lennewpopulation populationsize:
parent roulettewheelselectionpopulation fitnesses
parent roulettewheelselectionpopulation fitnesses
child child crossoverparent parent
newpopulation.extendmutatechild mutationrate mutatechild mutationrate
population newpopulation:populationsize
return maxpopulation keycalculatefitness
# Example graph as provided
graph
pu: pet
pe: putvg
v: gpe
g: petnv
n: gt
t: ngpepu
# Running the Genetic Algorithm
bestpath geneticalgorithmgraph populationsize generations
printBest Path:", bestpath
printFitness: calculatefitnessbestpath
Step by Step Solution
There are 3 Steps involved in it
1 Expert Approved Answer
Step: 1 Unlock


Question Has Been Solved by an Expert!
Get step-by-step solutions from verified subject matter experts
Step: 2 Unlock
Step: 3 Unlock


