Question: please give me the correct answer , if you want the singleFileData give me you email or any way to send the file Total of

please give me the correct answer , if you want the singleFileData give me you email or any way to send the file
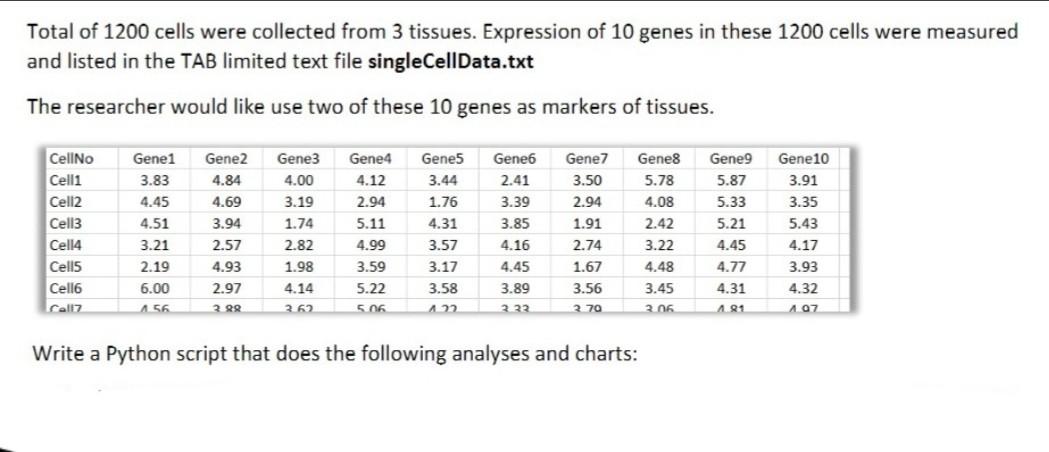
Total of 1200 cells were collected from 3 tissues. Expression of 10 genes in these 1200 cells were measured and listed in the TAB limited text file singleCellData.txt The researcher would like use two of these 10 genes as markers of tissues. CellNo Celli Cell2 Cell3 Cell4 Cells Cell6 call Gene1 3.83 4.45 4.51 3.21 2.19 6.00 156 Gene2 4.84 4.69 3.94 2.57 4.93 Gene3 4.00 3.19 1.74 2.82 1.98 Gene4 4.12 2.94 5.11 4.99 3.59 5.22 5.06 Gene5 3.44 1.76 4.31 3.57 3.17 3.58 122 Gene6 2.41 3.39 3.85 4.16 4.45 3.89 2.22 Gene7 3.50 2.94 1.91 2.74 1.67 3.56 2.70 Gene8 5.78 4.08 2.42 3.22 4.48 3.45 206 Gene9 5.87 5.33 5.21 4.45 4.77 4.31 181 Gene10 3.91 3.35 5.43 4.17 3.93 4.32 107 2.97 4.14 3.62 2. RR Write a Python script that does the following analyses and charts: 4. (15 points) Plot expression values of these two genes against each other (such as GeneX vs Genez) 5. (10 points) Decide on the threshold expression values of GeneX and GeneZ that will identify the origin of tissue for each cell. Write these values as comments in your code: # Upper and lower threshold of GeneX for Tissuel is chosen as XX.XX and YY. VY....etc 6. (15 points) Use the thresholds to classify each cell, is it from Tissue1, Tissue2 or Tissue3? If some cells cannot be identified, label them as unidentified
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


