Question: prefered. A simple model of this process can be given by the equations: d{cirne = pel = pero d[clprod] = wc[cIrna] Xel prot[cIprot] dt (1)
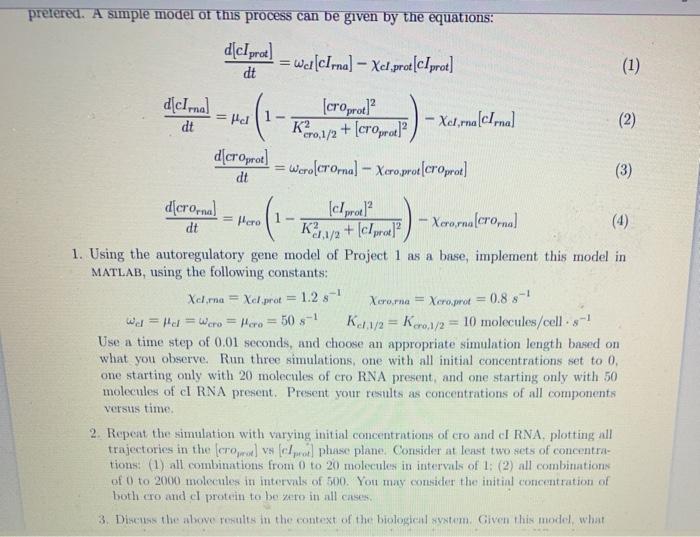
prefered. A simple model of this process can be given by the equations: d{cirne = pel = pero d[clprod] = wc[cIrna] Xel prot[cIprot] dt (1) [croprote] - Xara cima (2) dt K30,1/2 + (croprot) d[croprot) = Wero(crornal - Xero, prot[croprot] (3) dt d[crorna (c/prod? - Xorora (crorna (4) dt K3,1/2 + (cIprot? 1. Using the autoregulatory gene model of Project 1 as a base, implement this model in MATLAB, using the following constants: Xcima = Xel.prot = = 1.25-1 Xerorna = Xcroprot wd = Hal = wcro = Here = 50 s-1 Kc1/2 = Kr.1/2 = 10 molecules/cell.:-! Use a time step of 0.01 seconds, and choose an appropriate simulation length based on what you observe. Run three simulations, one with all initial concentrations set to 0. one starting only with 20 molecules of cro RNA present, and one starting only with 50 molecules of cl RNA present. Present your results as concentrations of all components versus time 2. Repeat the simulation with varying initial concentrations of cro and I RNA, plotting all trajectories in the cromo) vs ( coi phase plane. Consider at least two sets of concentra- tions: (1) all combinations from 0 to 20 molecules in intervals of 1: (2) all combinations of 0 to 2000 molecules in intervals of 500. You may consider the initind concentration of both cro and el protein to be vero in all cases 3. Discuss the above results in the context of the biological system. Given this model, what 0.8 -1
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


