Question: Question 4: Use the protein sequence alignment to answer the questions. The sequences belong to the same domains of the S1=Progesterone Receptor, S2=Androgen Receptor, S3=Glucocorticoid
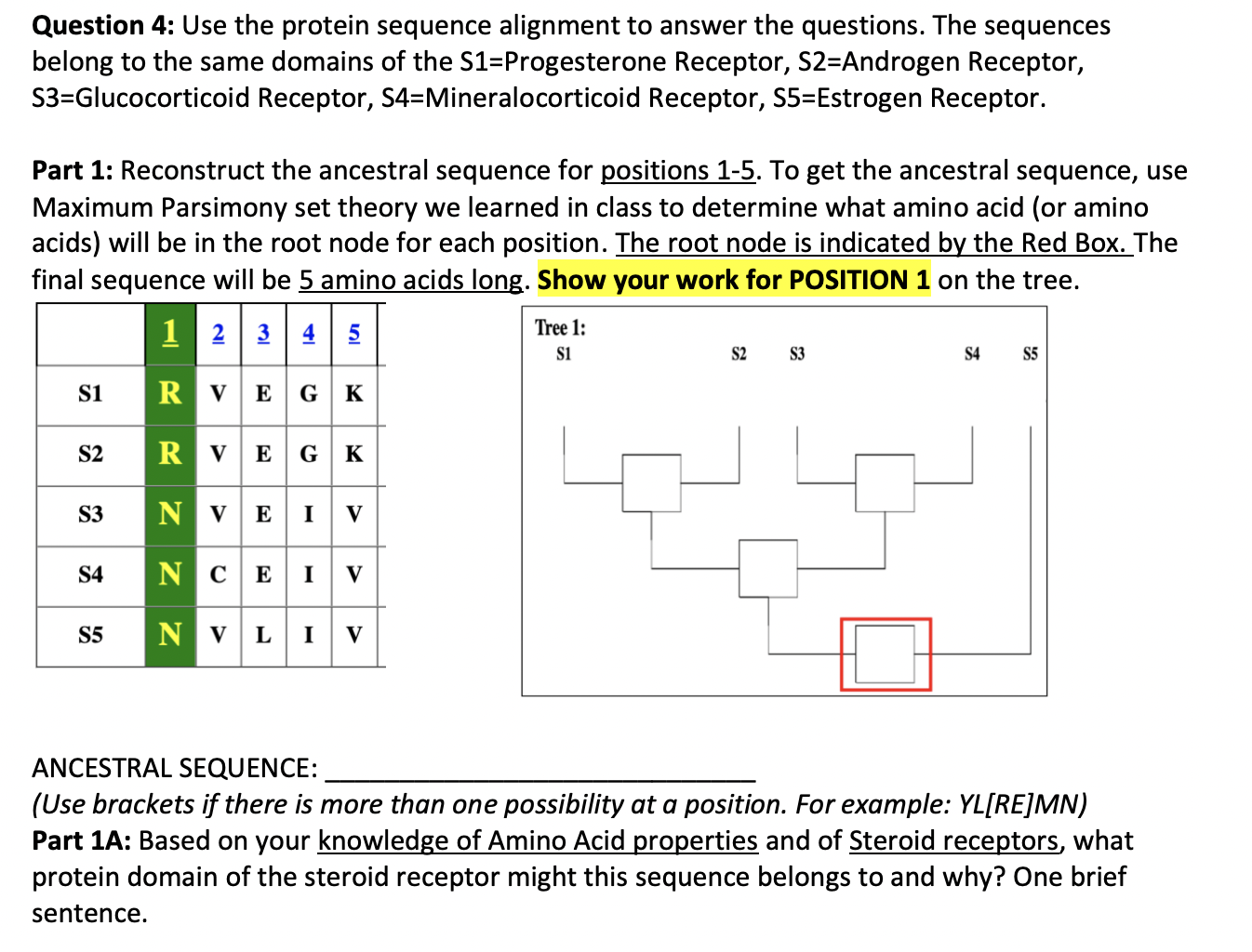
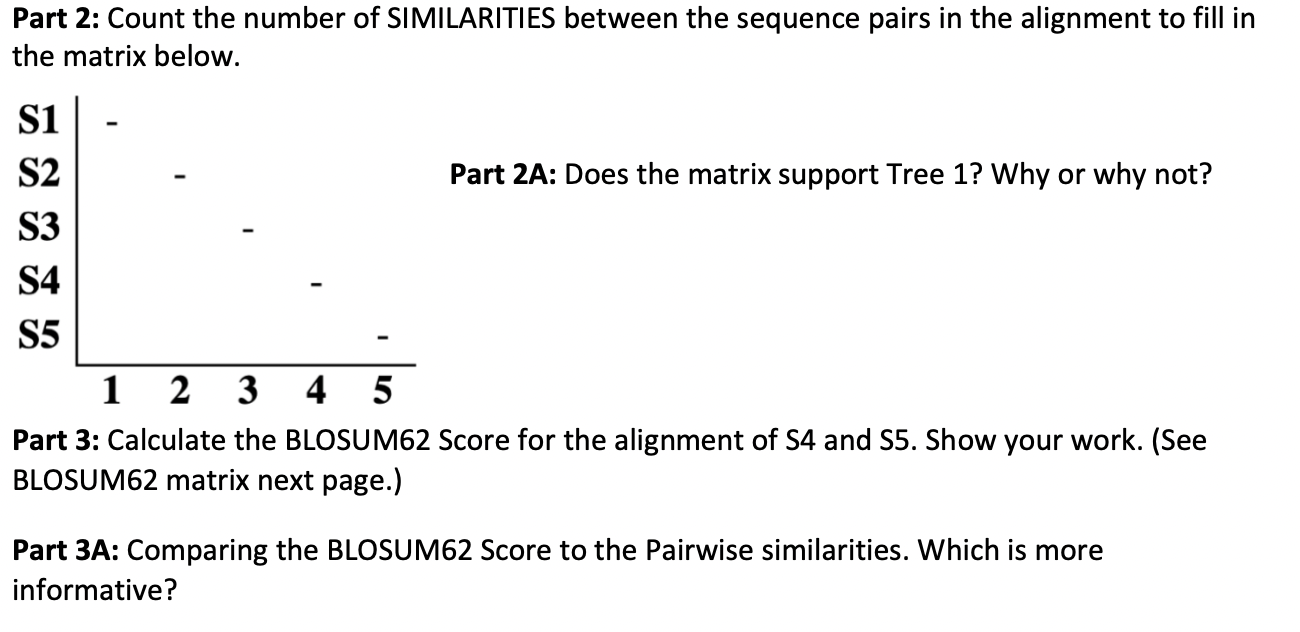
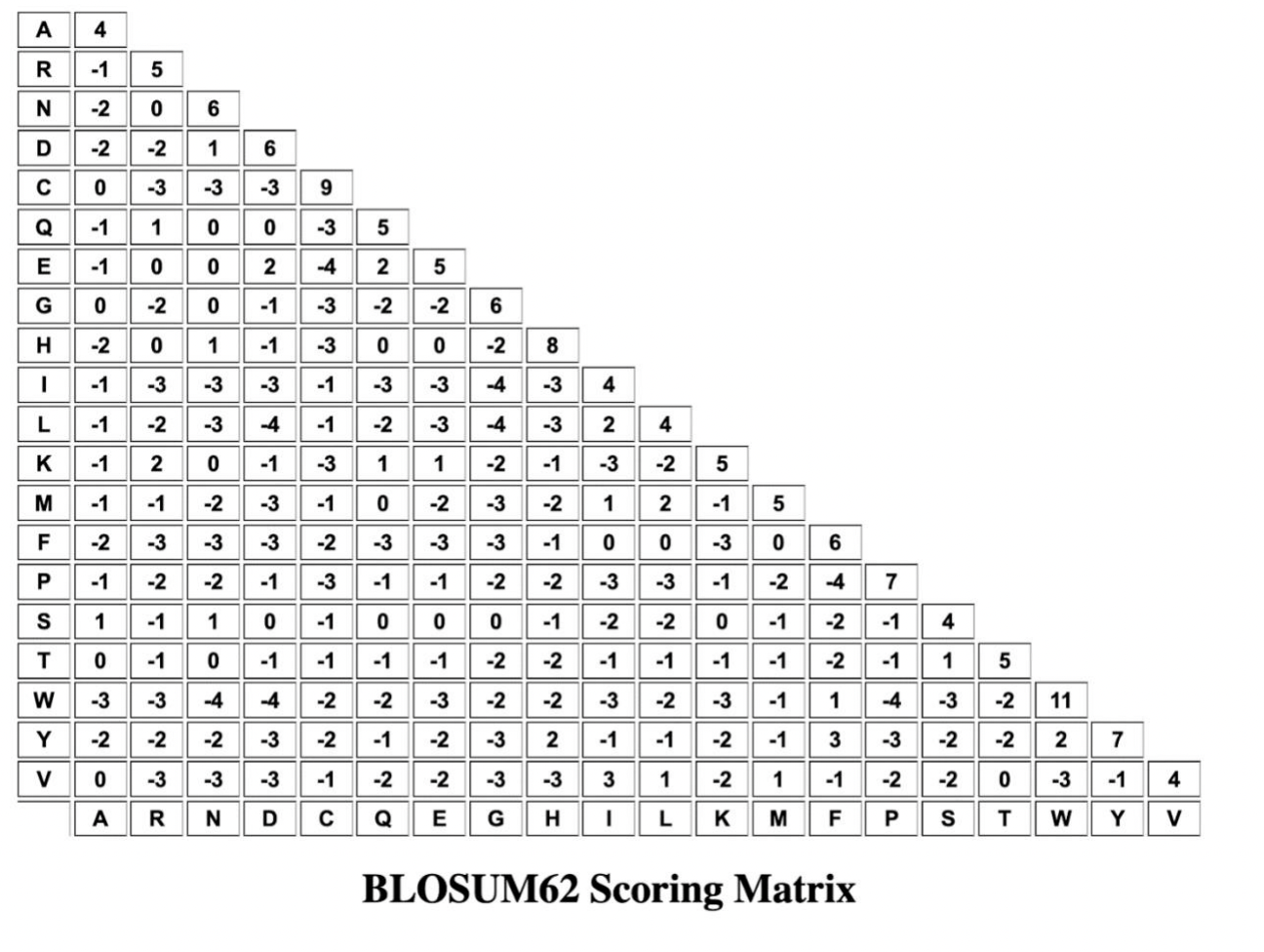
Question 4: Use the protein sequence alignment to answer the questions. The sequences belong to the same domains of the S1=Progesterone Receptor, S2=Androgen Receptor, S3=Glucocorticoid Receptor, S4=Mineralocorticoid Receptor, S5=Estrogen Receptor. Part 1: Reconstruct the ancestral sequence for positions 1-5. To get the ancestral sequence, use Maximum Parsimony set theory we learned in class to determine what amino acid (or amino acids) will be in the root node for each position. The root node is indicated by the Red Box. The final sequence will be 5 amino acids long. Show your work for POSITION 1 on the tree. 1 2 | 3 | 4 | 5 Tree 1: si S2 S3 S4 S5 S1 R V E G K S2 R R ve GK S3 N v E I V S4 N c E 1 v S5 N V L I V ANCESTRAL SEQUENCE: (Use brackets if there is more than one possibility at a position. For example: YL[RE]MN) Part 1A: Based on your knowledge of Amino Acid properties and of Steroid receptors, what protein domain of the steroid receptor might this sequence belongs to and why? One brief sentence. Part 2: Count the number of SIMILARITIES between the sequence pairs in the alignment to fill in the matrix below. S1 S2 Part 2A: Does the matrix support Tree 1? Why or why not? S3 S4 S5 1 2 3 4 5 Part 3: Calculate the BLOSUM62 Score for the alignment of S4 and S5. Show your work. (See BLOSUM62 matrix next page.) - Part 3A: Comparing the BLOSUM62 Score to the Pairwise similarities. Which is more informative? 4 G -3 -3 -3 -3 4 R -1 5 N -2 0 6 D -2 -2 16 0 -3 -3 -3 9 Q 1 0 0 -3 5 E 1 0 0 2 4 2 5 0 -2 0 -1 -3 -2 -2 6 -2 0 1 -1 -3 0 0 -2 8 -1 -3 -3 -1 -3 4 4 -1 -2 -4 -1 -2 -3 -4 -3 2 -1 2 0 -1 -3 1 1 -2 -1 -2 5 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 -2 -3 -3 -3 -2 -3 -3 -3 0 0 -3 0 6 0 -1-2 -2 -1 -3-1 -2 -2 -3 -3 -1 -2 4 7 s 1 -1 1 0 -1 0 0 0 -1 -2 -2 0-1 0 -2 -1 4 T 0 -1 0 -1 -1 -1 -2 -2 -1 -1 -1 -2 -1 1 5 w -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 4 -3 -2 11 -2 -2 -2 -3 -2 -2 -3 2 -1 -1 -2 -1 3 -2 -2 2 7 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1-1 3 -2 0 -3 -1 R N Q E G HT L K M F P T W Y 2 . -3 1-1-YELMES >> -3 -2 4 A D S V BLOSUM62 Scoring Matrix Question 4: Use the protein sequence alignment to answer the questions. The sequences belong to the same domains of the S1=Progesterone Receptor, S2=Androgen Receptor, S3=Glucocorticoid Receptor, S4=Mineralocorticoid Receptor, S5=Estrogen Receptor. Part 1: Reconstruct the ancestral sequence for positions 1-5. To get the ancestral sequence, use Maximum Parsimony set theory we learned in class to determine what amino acid (or amino acids) will be in the root node for each position. The root node is indicated by the Red Box. The final sequence will be 5 amino acids long. Show your work for POSITION 1 on the tree. 1 2 | 3 | 4 | 5 Tree 1: si S2 S3 S4 S5 S1 R V E G K S2 R R ve GK S3 N v E I V S4 N c E 1 v S5 N V L I V ANCESTRAL SEQUENCE: (Use brackets if there is more than one possibility at a position. For example: YL[RE]MN) Part 1A: Based on your knowledge of Amino Acid properties and of Steroid receptors, what protein domain of the steroid receptor might this sequence belongs to and why? One brief sentence. Part 2: Count the number of SIMILARITIES between the sequence pairs in the alignment to fill in the matrix below. S1 S2 Part 2A: Does the matrix support Tree 1? Why or why not? S3 S4 S5 1 2 3 4 5 Part 3: Calculate the BLOSUM62 Score for the alignment of S4 and S5. Show your work. (See BLOSUM62 matrix next page.) - Part 3A: Comparing the BLOSUM62 Score to the Pairwise similarities. Which is more informative? 4 G -3 -3 -3 -3 4 R -1 5 N -2 0 6 D -2 -2 16 0 -3 -3 -3 9 Q 1 0 0 -3 5 E 1 0 0 2 4 2 5 0 -2 0 -1 -3 -2 -2 6 -2 0 1 -1 -3 0 0 -2 8 -1 -3 -3 -1 -3 4 4 -1 -2 -4 -1 -2 -3 -4 -3 2 -1 2 0 -1 -3 1 1 -2 -1 -2 5 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 -2 -3 -3 -3 -2 -3 -3 -3 0 0 -3 0 6 0 -1-2 -2 -1 -3-1 -2 -2 -3 -3 -1 -2 4 7 s 1 -1 1 0 -1 0 0 0 -1 -2 -2 0-1 0 -2 -1 4 T 0 -1 0 -1 -1 -1 -2 -2 -1 -1 -1 -2 -1 1 5 w -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 4 -3 -2 11 -2 -2 -2 -3 -2 -2 -3 2 -1 -1 -2 -1 3 -2 -2 2 7 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1-1 3 -2 0 -3 -1 R N Q E G HT L K M F P T W Y 2 . -3 1-1-YELMES >> -3 -2 4 A D S V BLOSUM62 Scoring Matrix
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


