Question: Show the theoretical time complexity of the Sequence Alignment Divide and Conquer algorithm. For the recursive algorithm change the recurrence relation to act as if
Show the theoretical time complexity of the Sequence Alignment Divide and Conquer algorithm. For the recursive algorithm change the recurrence relation to act as if it was one variable. Start from the algorithm and develop the time complexity, then simplify the problem to get a problem with one variable, and obtain a pseudo recurrence relation using only one variable.
Algorithm:
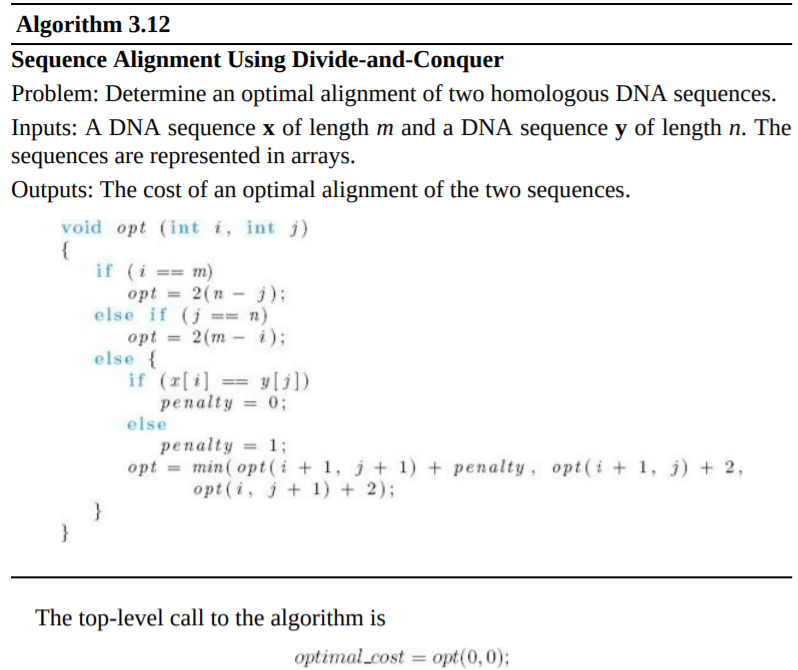
Algorithm 3.12 Sequence Alignment Using Divide-and-Conquer Problem: Determine an optimal alignment of two homologous DNA sequences. Inputs: A DNA sequence x of length m and a DNA sequence y of length n. The sequences are represented in arrays. Outputs: The cost of an optimal alignment of the two sequences. void opt (int i, inti) { if (i == m) opt = 2(n - i); else if (j == n) opt 2(m - i); else { if (x[i] == y(1) penalty = 0; else penalty = 1; opt = min( opt(i+1, j + 1) + penalty, opt(i+1, j) + 2, opt(i, j + 1) + 2); The top-level call to the algorithm is optimal_cost = opt(0, 0)
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


