Question: Your program is responsible for performing only the single mutation that occurs between the first occurrence of the pattern, and the next subsequent occurrence of
Your program is responsible for performing only the single mutation that occurs between the first occurrence of the pattern, and the next subsequent occurrence of the reversed pattern (that which is completely disjoint from the forward pattern).
However, you need not concern yourself with how your program behaves if given errant input that does not meet these formal specifications. (In a later chapter, we will learn about conditional statements which would allow us to gracefully handle such situations.)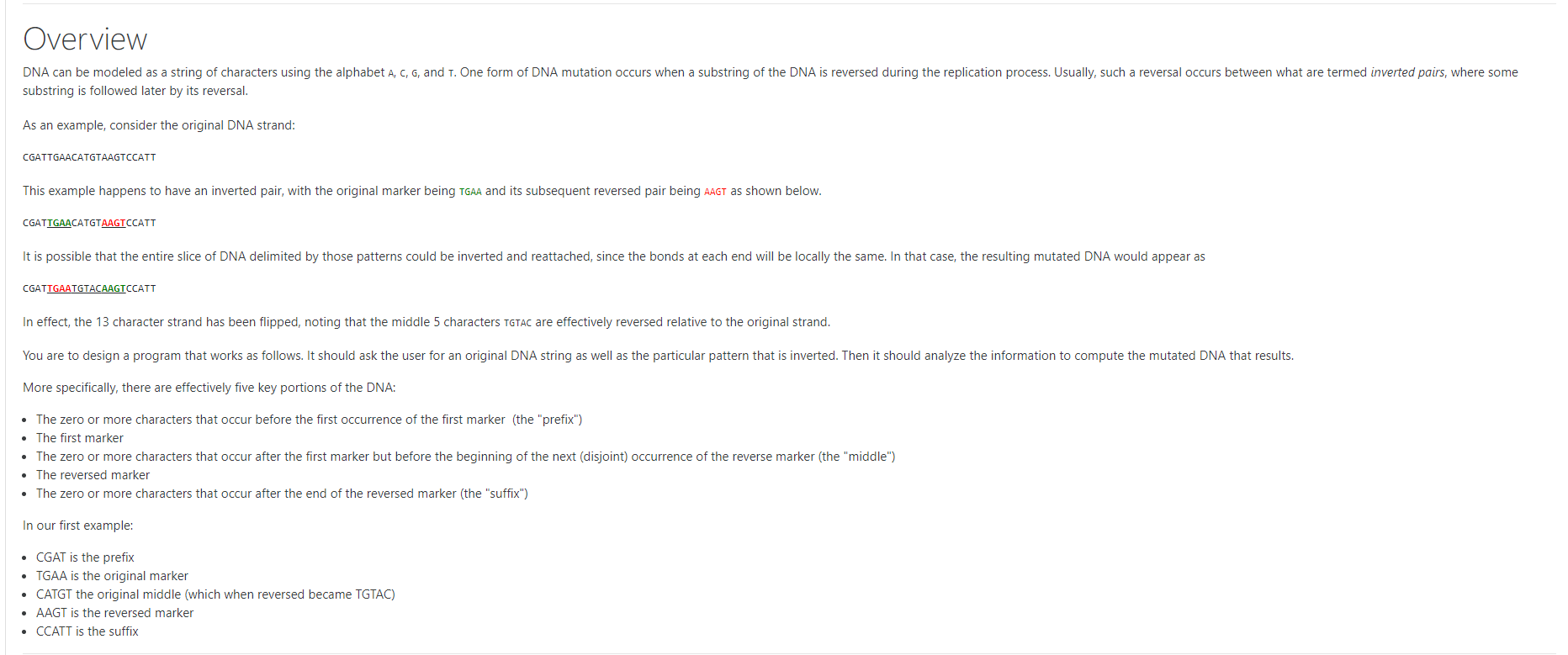
Note: I need help solving this project via Python, and also it cannnot be done using loops. Thank You.
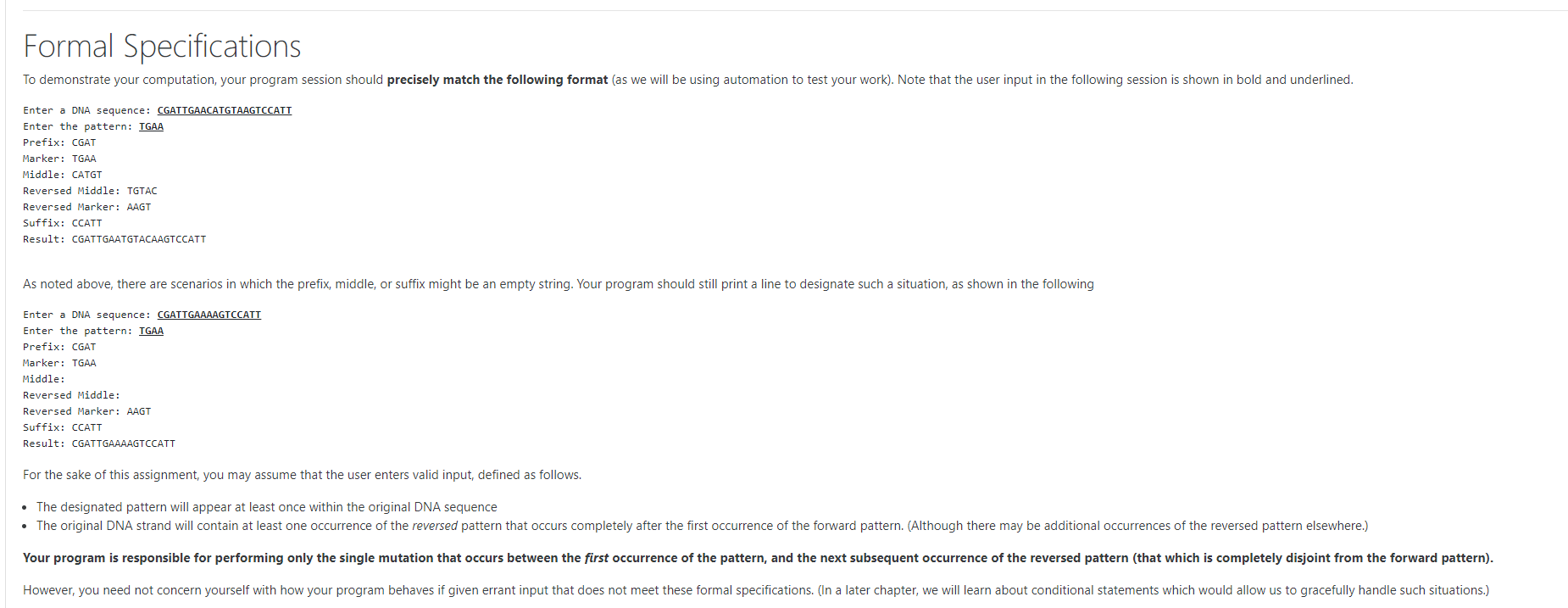
Overview DNA can be modeled as a string of characters using the alphabet A, C, G, and 1. One form of DNA mutation occurs when a substring of the DNA is reversed during the replication process. Usually, such a reversal occurs between what are termed inverted pairs, where some substring is followed later by its reversal. As an example, consider the original DNA strand: CGATTGAACATGTAAGTCCATT This example happens to have an inverted pair, with the original marker being TGA and its subsequent reversed pair being AAGT as shown below. CGATTGAACATGTAAGTCCATT It is possible that the entire slice of DNA delimited by those patterns could be inverted and reattached, since the bonds at each end will be locally the same. In that case, the resulting mutated DNA would appear as CGATTGAATGTACAAGTCCATT In effect, the 13 character strand has been flipped, noting that the middle 5 characters TGTAC are effectively reversed relative to the original strand. You are to design a program that works as follows. It should ask the user for an original DNA string as well as the particular pattern that is inverted. Then it should analyze the information to compute the mutated DNA that results. More specifically, there are effectively five key portions of the DNA: The zero or more characters that occur before the first occurrence of the first marker (the "prefix") The first marker The zero or more characters that occur after the first marker but before the beginning of the next (disjoint) occurrence of the reverse marker (the "middle") The reversed marker The zero or more characters that occur after the end of the reversed marker (the "suffix") In our first example: CGAT is the prefix TGAA is the original marker CATGT the original middle (which when reversed became TGTAC) AAGT is the reversed marker CCATT is the suffix Formal Specifications To demonstrate your computation, your program session should precisely match the following format (as we will be using automation to test your work). Note that the user input in the following session is shown in bold and underlined. Enter a DNA sequence: CGATTGAACATGTAAGTCCATT Enter the pattern: TGAA Prefix: CGAT Marker: TGAA Middle: CATGT Reversed Middle: TGTAC Reversed Marker: AAGT Suffix: CCATT Result: CGATTGAATGTACAAGTCCATT As noted above, there are scenarios in which the prefix, middle, or suffix might be an empty string. Your program should still print a line to designate such a situation, as shown in the following Enter a DNA sequence: CGATTGAAAAGTCCATT Enter the pattern: TGAA Prefix: CGAT Marker: TGAA Middle: Reversed Middle: Reversed Marker: AAGT Suffix: CCATT Result: CGATTGAAAAGTCCATT For the sake of this assignment, you may assume that the user enters valid input, defined as follows. The designated pattern will appear at least once within the original DNA sequence The original DNA strand will contain at least one occurrence of the reversed pattern that occurs completely after the first occurrence of the forward pattern. (Although there may be additional occurrences of the reversed pattern elsewhere.) Your program is responsible for performing only the single mutation that occurs between the first occurrence of the pattern, and the next subsequent occurrence of the reversed pattern (that which is completely disjoint from the forward pattern). However, you need not concern yourself with how your program behaves if given errant input that does not meet these formal specifications. (In a later chapter, we will learn about conditional statements which would allow us to gracefully handle such situations.)
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


