Question: 1. Regression with Polynomial Basis Functions, 30 points. This problem extends ordinary least squares regression, which uses the hypothesis class of linear regression functions, to
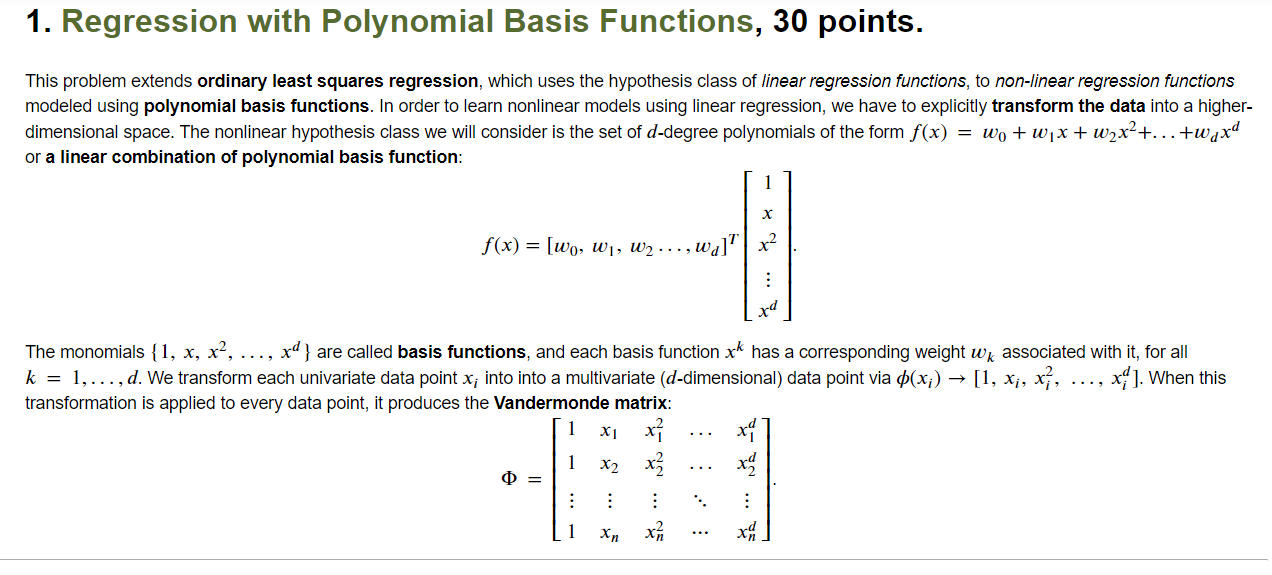
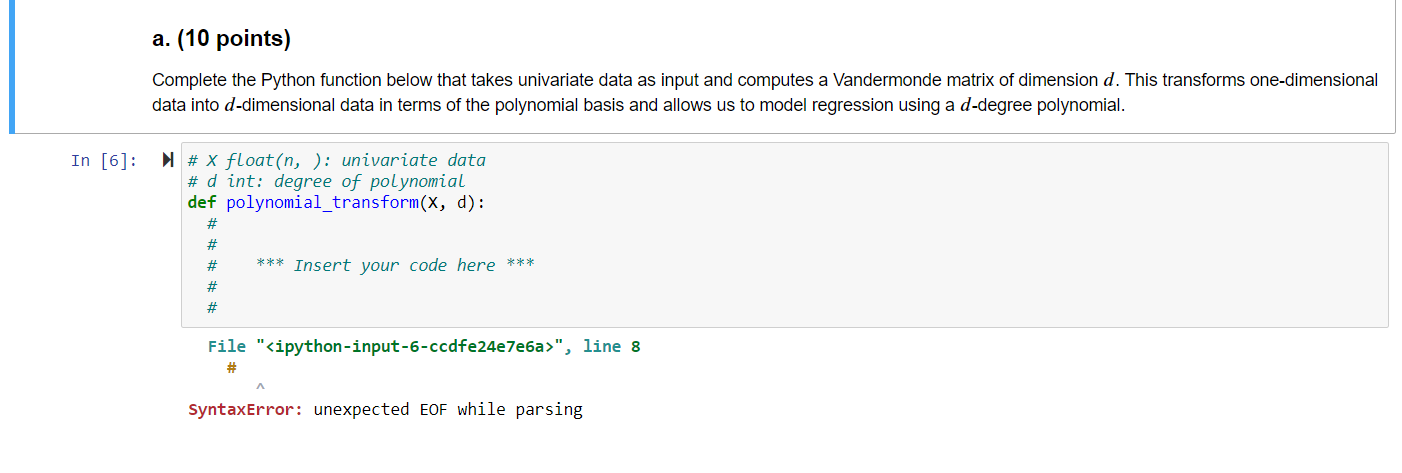
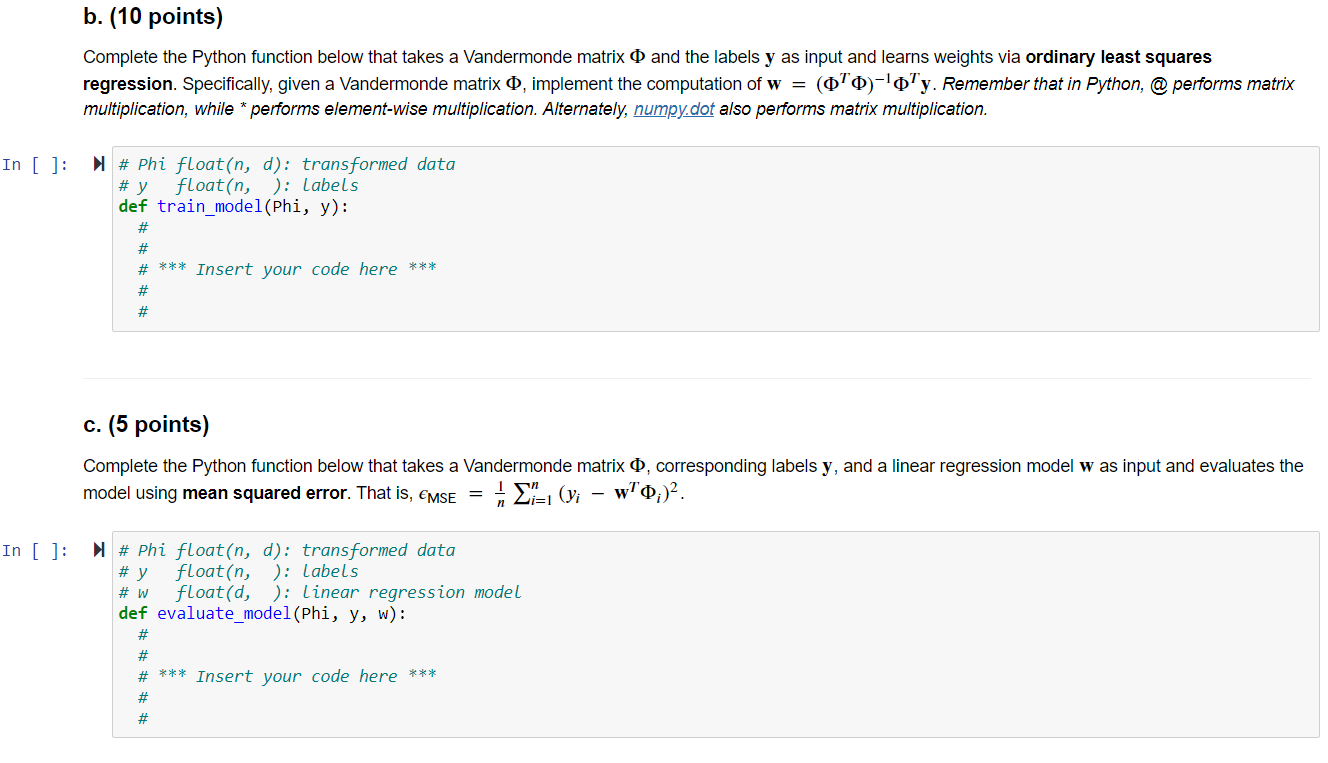
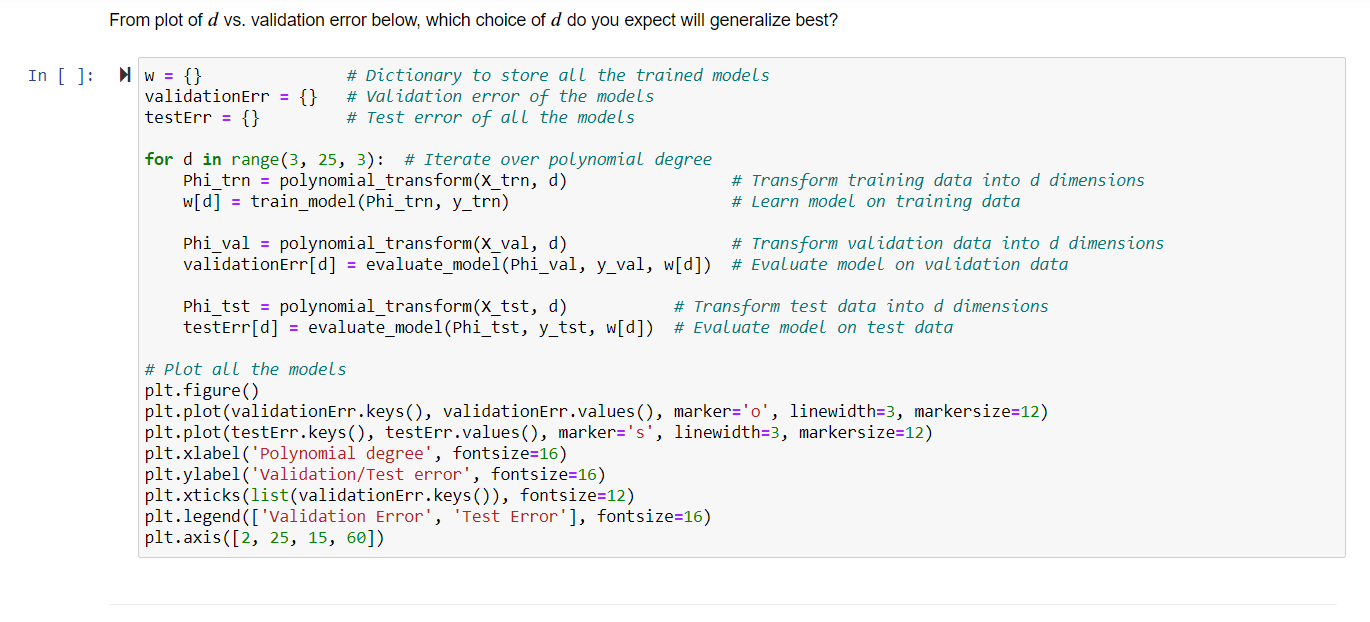
1. Regression with Polynomial Basis Functions, 30 points. This problem extends ordinary least squares regression, which uses the hypothesis class of linear regression functions, to non-linear regression functions modeled using polynomial basis functions. In order to learn nonlinear models using linear regression, we have to explicitly transform the data into a higher- dimensional space. The nonlinear hypothesis class we will consider is the set of d-degree polynomials of the form f(x) = wo + w1x + w2x+...+wqxd or a linear combination of polynomial basis function: X f(x) = [wo, W1, W2 ..., wa] x2 rd The monomials {1, x, x, ..., xd } are called basis functions, and each basis function xk has a corresponding weight wk associated with it, for all k = 1,..., d. We transform each univariate data point x; into into a multivariate (d-dimensional) data point via p(x;) [1, x, xz, x]. When this transformation is applied to every data point, it produces the Vandermonde matrix: X1 x? 1 x2 x? : : 0 = 1 xn a. (10 points) Complete the Python function below that takes univariate data as input and computes a Vandermonde matrix of dimension d. This transforms one-dimensional data into d-dimensional data in terms of the polynomial basis and allows us to model regression using a d-degree polynomial. In [6]: # x float(n, ): univariate data # d int: degree of polynomial def polynomial_transform(x, d): # # # *** Insert your code here *** # # File "", line 8 SyntaxError: unexpected EOF while parsing b. (10 points) Complete the Python function below that takes a Vandermonde matrix o and the labels y as input and learns weights via ordinary least squares regression. Specifically, given a Vandermonde matrix O, implement the computation of w = (070)-'0'y. Remember that in Python, @performs matrix multiplication, while * performs element-wise multiplication. Alternately, numpy.dot also performs matrix multiplication. In [ ]: # Phi float(n, d): transformed data # y float(n, ): Labels def train_model(Phi, y): # # # *** Insert your code here *** # # c. (5 points) Complete the Python function below that takes a Vandermonde matrix o, corresponding labels y, and a linear regression model w as input and evaluates the model using mean squared error. That is, emse = X= (yi wo;) In [ ]: # Phi float(n, d): transformed data # y float(n, ): Labels float(d, ): Linear regression model def evaluate_model(Phi, y, w): # w # # # *** Insert your code here *** # # From plot of d vs. validation error below, which choice of d do you expect will generalize best? In [ ]: W = {} validationErr = testErr = {} {} # Dictionary to store all the trained models # Validation error of the models # Test error of all the models for d in range(3, 25, 3): # Iterate over polynomial degree Phi_trn = polynomial_transform(x_trn, d) w[d] = train_model(Phi_trn, y tnn) # Transform training data into d dimensions # Learn model on training data Phi_val = polynomial_transform(X_val, d) # Transform validation data into d dimensions validationErr[d] = evaluate_model(Phi_val, y_val, w[d]) # Evaluate model on validation data Phi_tst = polynomial_transform(x_tst, d) # Transform test data into d dimensions testErr[d] = evaluate_model(Phi_tst, y_tst, w[d]) # Evaluate model on test data # Plot all the models plt.figure() plt.plot(validationErr.keys(), validationErr.values(), marker='o', linewidth=3, markersize=12) plt.plot(testErr.keys(), testErr.values(), marker='s', linewidth=3, markersize=12) plt.xlabel('Polynomial degree', fontsize=16) plt.ylabel('Validation/Test error', fontsize=16) plt.xticks (list(validationErr.keys()), fontsize=12) plt.legend (['Validation Error', 'Test Error'], fontsize=16) plt.axis ([2, 25, 15, 60]) 1. Regression with Polynomial Basis Functions, 30 points. This problem extends ordinary least squares regression, which uses the hypothesis class of linear regression functions, to non-linear regression functions modeled using polynomial basis functions. In order to learn nonlinear models using linear regression, we have to explicitly transform the data into a higher- dimensional space. The nonlinear hypothesis class we will consider is the set of d-degree polynomials of the form f(x) = wo + w1x + w2x+...+wqxd or a linear combination of polynomial basis function: X f(x) = [wo, W1, W2 ..., wa] x2 rd The monomials {1, x, x, ..., xd } are called basis functions, and each basis function xk has a corresponding weight wk associated with it, for all k = 1,..., d. We transform each univariate data point x; into into a multivariate (d-dimensional) data point via p(x;) [1, x, xz, x]. When this transformation is applied to every data point, it produces the Vandermonde matrix: X1 x? 1 x2 x? : : 0 = 1 xn a. (10 points) Complete the Python function below that takes univariate data as input and computes a Vandermonde matrix of dimension d. This transforms one-dimensional data into d-dimensional data in terms of the polynomial basis and allows us to model regression using a d-degree polynomial. In [6]: # x float(n, ): univariate data # d int: degree of polynomial def polynomial_transform(x, d): # # # *** Insert your code here *** # # File "", line 8 SyntaxError: unexpected EOF while parsing b. (10 points) Complete the Python function below that takes a Vandermonde matrix o and the labels y as input and learns weights via ordinary least squares regression. Specifically, given a Vandermonde matrix O, implement the computation of w = (070)-'0'y. Remember that in Python, @performs matrix multiplication, while * performs element-wise multiplication. Alternately, numpy.dot also performs matrix multiplication. In [ ]: # Phi float(n, d): transformed data # y float(n, ): Labels def train_model(Phi, y): # # # *** Insert your code here *** # # c. (5 points) Complete the Python function below that takes a Vandermonde matrix o, corresponding labels y, and a linear regression model w as input and evaluates the model using mean squared error. That is, emse = X= (yi wo;) In [ ]: # Phi float(n, d): transformed data # y float(n, ): Labels float(d, ): Linear regression model def evaluate_model(Phi, y, w): # w # # # *** Insert your code here *** # # From plot of d vs. validation error below, which choice of d do you expect will generalize best? In [ ]: W = {} validationErr = testErr = {} {} # Dictionary to store all the trained models # Validation error of the models # Test error of all the models for d in range(3, 25, 3): # Iterate over polynomial degree Phi_trn = polynomial_transform(x_trn, d) w[d] = train_model(Phi_trn, y tnn) # Transform training data into d dimensions # Learn model on training data Phi_val = polynomial_transform(X_val, d) # Transform validation data into d dimensions validationErr[d] = evaluate_model(Phi_val, y_val, w[d]) # Evaluate model on validation data Phi_tst = polynomial_transform(x_tst, d) # Transform test data into d dimensions testErr[d] = evaluate_model(Phi_tst, y_tst, w[d]) # Evaluate model on test data # Plot all the models plt.figure() plt.plot(validationErr.keys(), validationErr.values(), marker='o', linewidth=3, markersize=12) plt.plot(testErr.keys(), testErr.values(), marker='s', linewidth=3, markersize=12) plt.xlabel('Polynomial degree', fontsize=16) plt.ylabel('Validation/Test error', fontsize=16) plt.xticks (list(validationErr.keys()), fontsize=12) plt.legend (['Validation Error', 'Test Error'], fontsize=16) plt.axis ([2, 25, 15, 60])







