Question: 3 reading frames EDITED - The script that I attached below needs to be modified so that it returns a list of 3 amino acid
3 reading frames
EDITED - The script that I attached below needs to be modified so that it returns a list of 3 amino acid strings. A function (forloop) that can slice the DNA into three different reading frames and translate them as three different among acids. And then returns the amino acid string three times. the question below has examples of what it should return.
in different words, if I input a DNA string that is ATGCTCGTATAAGGGCCATTGCG, then my function should start reading the first frame staring at ATG example amino_acid_1 = translate(dan[0:1]). So entire dan string that starts at first letter.
Then the second frame from the next letter, TGC example amino_acid_2 = translate(dan[1:]). So entire DNA string starting at second alphabet
Then third frame from third letter, GCT example amino_acid_3 = translate(dan[2:]). So entire DNA string starting at third alphabet.
-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -- -
hi. I already have a python script (picture below) that takes a protein script, translates it and gives me back amino acid. (this info was imported from a table). I need to just MODIFY this script. (I have included example below)
I need to use a FOR LOOP that is programmed to SLICE the DNA (three letters at a time), CALLS the translate function three times that RETURNS amino acid string. Then that returned function should APPEND to a LIST of reading frames.
so at the end we will have a list of three reading frames, each one is a string of amino acids. 
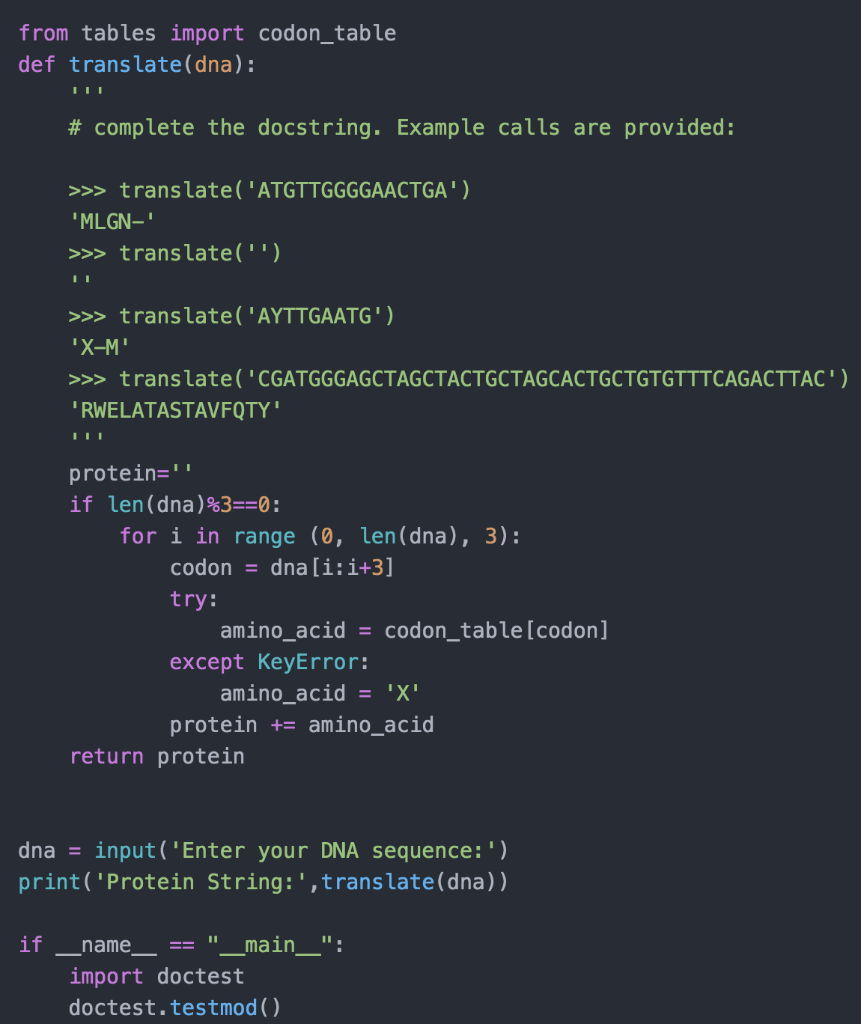
Write a new function that: 1. takes a single input parameter, a dna string 2. slices the dna string into 3 reading frames 3. calls your existing translate function 3 times, once for each of the 3 reading frames 4. stores 3 amino acid strings in a list. Your translate function will return an amino acid string 3 times. Store all 3. 5. returns that list INPUT: a dna string RETURN: a list of three amino acid strings e.g. This DNA string: dna = 'ATGTTGGGGAACTGA' Can be translated into these 3 amino-acid sequences. 'MLGN-' 'CWGT' 'VGEL' So the returned value is this list: ['MLGN-' CWGT', 'VGEL'] from tables import codon_table def translate(dna): # complete the docstring. Example calls are provided : >>> translate('ATGTTGGGGAACTGA'). "MLGN-' >>> translate("') >>> translate('AYTTGAATG') "X-M' >>> translate('CGATGGGAGCTAGCTACTGCTAGCACTGCTGTGTTTCAGACTTAC') "RWELATASTAVFQTY' protein=" if len(dna)%3==0: for i in range (0, len(dna), 3): codon = dna[i:i+3] try: amino_acid = codon_table(codon] except KeyError: amino acid = 'X' protein += amino acid return protein dna = input('Enter your DNA sequence:') print('Protein String:', translate(dna)) if name _main__": import doctest doctest.testmod()
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


