Question: Consider the Sequence-Alignment problem on the DNA sequences CGAAGTCA and CTCAAGA using a gap penalty of =1 and the substitution matrix: For each of the
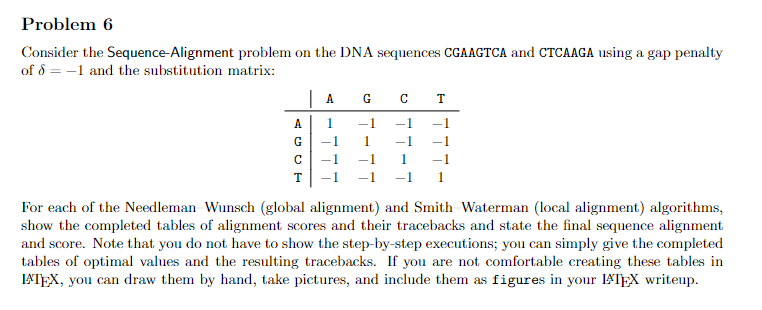
Consider the Sequence-Alignment problem on the DNA sequences CGAAGTCA and CTCAAGA using a gap penalty of =1 and the substitution matrix: For each of the Needleman Wunsch (global alignment) and Smith Waterman (local alignment) algorithms, show the completed tables of alignment scores and their tracebacks and state the final sequence alignment and score. Note that you do not have to show the step-by-step executions; you can simply give the completed tables of optimal values and the resulting tracebacks. If you are not comfortable creating these tables in ATEX, you can draw them by hand, take pictures, and include them as figures in your ETEX writeup
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


