Question: here is the link for the csv file http://www.dewetcomputers.com/CSV/dataset_assignment3.csv The project asks you to develop, evaluate and compare models for the prediction of proteins that

here is the link for the csv file
http://www.dewetcomputers.com/CSV/dataset_assignment3.csv
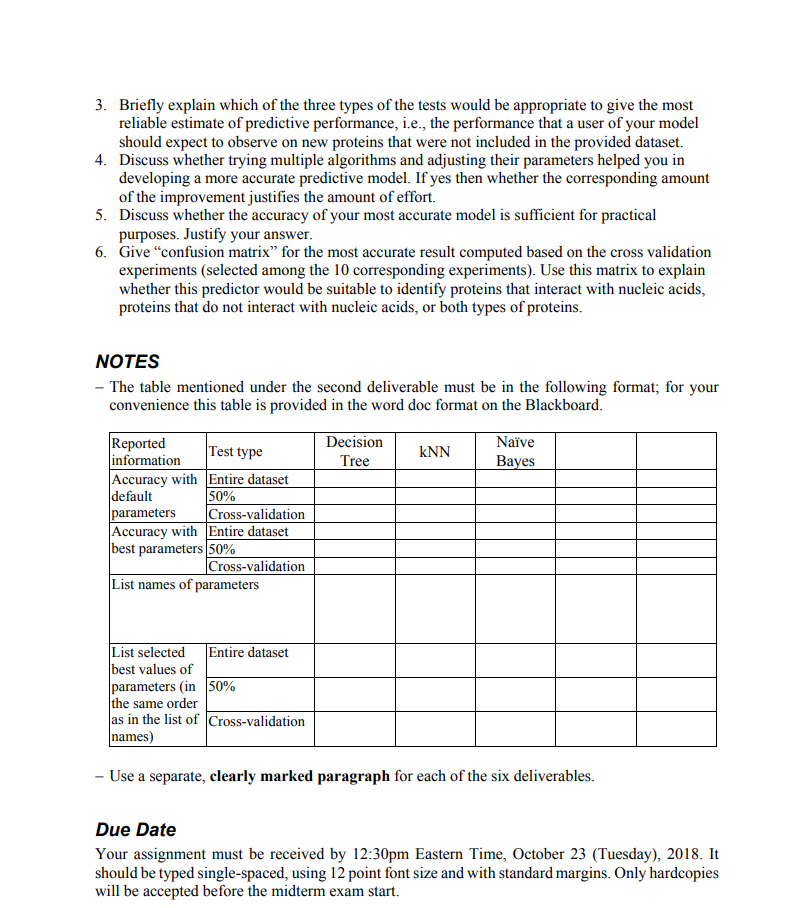
The project asks you to develop, evaluate and compare models for the prediction of proteins that interact with nucleic acids (either DNA or RNA) using a provided dataset. Dataset The dataset ("dataset assignment3.csv" file) is provided in the text-based, comma-separated format where each protein is represented by 13 numeric features and 1 symbolic outcome. The outcome feature (called "class") annotates each proteins as Y (interacting) vs. N (non- interacting). The dataset includes 8795 proteins, with 936 labeled Y (interacting with nucleic acids) and 7859 labeled N (not interacting with nucleic acids) Development of predictive models You are required to compute models with version 9.0 of the RapidMiner Studio using five different algorithms. Three of these five algorithms must be the Decision Tree, kNN and Nave Bayes. You can choose any of the other predictive algorithms for the remaining two. You shoulod parametrize each of these algorithms (select the best possible combination of values of their parameters), to the best of your ability, in order to maximize predictive performance. Note that you will need to read, make an educated guess, and/or use trial-and-error approach to figure out which parameters make a difference and how to use them. Do not use the "advanced parameters". Do not attempt to sample the dataset, i.e., do not perform feature or sample/object selection. Evaluation and comparison of predictive models You must evaluate the predictive performance using accuracy (1% of correctly classified instances"). For each algorithm you must perform three types of tests on the entire dataset (use training dataset") on 50% of the dataset; you will use the other 50% to compute the model ("percentage split") - using the 10 fold cross-validation The 10 fold cross-validation divides the dataset at random into 10 equal-size subsets, where one subset is used to test the model and the remaining nine to compute the prediction model. This is repeated 10 times, each time using a different subset as the test set. Consequently, this test results in predicting every protein in the dataset. This test type is implemented in the RapidMiner Studio with the "Cross Validation" operator where the number of folds is set to 10 1. List and briefly describe the methods that you used (one sentence per method) and list their ey parameters Using the table shown below, report the accuracies for the five algorithms and the three test types. The accuracy values must be reported with two digits after the decimal point, e.g., 91.05. You must include the accuracies of the models that use default parameters and the best selected parameters. In total, you have 5*3*2 = 30 results to report. List the best selected values of parameters for each model and each test type 2. The project asks you to develop, evaluate and compare models for the prediction of proteins that interact with nucleic acids (either DNA or RNA) using a provided dataset. Dataset The dataset ("dataset assignment3.csv" file) is provided in the text-based, comma-separated format where each protein is represented by 13 numeric features and 1 symbolic outcome. The outcome feature (called "class") annotates each proteins as Y (interacting) vs. N (non- interacting). The dataset includes 8795 proteins, with 936 labeled Y (interacting with nucleic acids) and 7859 labeled N (not interacting with nucleic acids) Development of predictive models You are required to compute models with version 9.0 of the RapidMiner Studio using five different algorithms. Three of these five algorithms must be the Decision Tree, kNN and Nave Bayes. You can choose any of the other predictive algorithms for the remaining two. You shoulod parametrize each of these algorithms (select the best possible combination of values of their parameters), to the best of your ability, in order to maximize predictive performance. Note that you will need to read, make an educated guess, and/or use trial-and-error approach to figure out which parameters make a difference and how to use them. Do not use the "advanced parameters". Do not attempt to sample the dataset, i.e., do not perform feature or sample/object selection. Evaluation and comparison of predictive models You must evaluate the predictive performance using accuracy (1% of correctly classified instances"). For each algorithm you must perform three types of tests on the entire dataset (use training dataset") on 50% of the dataset; you will use the other 50% to compute the model ("percentage split") - using the 10 fold cross-validation The 10 fold cross-validation divides the dataset at random into 10 equal-size subsets, where one subset is used to test the model and the remaining nine to compute the prediction model. This is repeated 10 times, each time using a different subset as the test set. Consequently, this test results in predicting every protein in the dataset. This test type is implemented in the RapidMiner Studio with the "Cross Validation" operator where the number of folds is set to 10 1. List and briefly describe the methods that you used (one sentence per method) and list their ey parameters Using the table shown below, report the accuracies for the five algorithms and the three test types. The accuracy values must be reported with two digits after the decimal point, e.g., 91.05. You must include the accuracies of the models that use default parameters and the best selected parameters. In total, you have 5*3*2 = 30 results to report. List the best selected values of parameters for each model and each test type 2
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


