Question: I have included the code for the .h and unit tester .cpp. C++ language is preferred. //Bring in unit testing code and tell it to
I have included the code for the .h and unit tester .cpp. C++ language is preferred. //Bring in unit testing code and tell it to build a main function
#define DOCTEST_CONFIG_IMPLEMENT_WITH_MAIN
//This pragma supresses a bunch of warnings QTCreator produces (and should not)
#pragma clang diagnostic ignored "-Woverloaded-shift-op-parentheses"
#include "doctest.h"
//Use Approx from doctest without saying doctest::Approx
using doctest::Approx;
#include
//Include your .h files
#include "DNAStrand.h"
using namespace std;
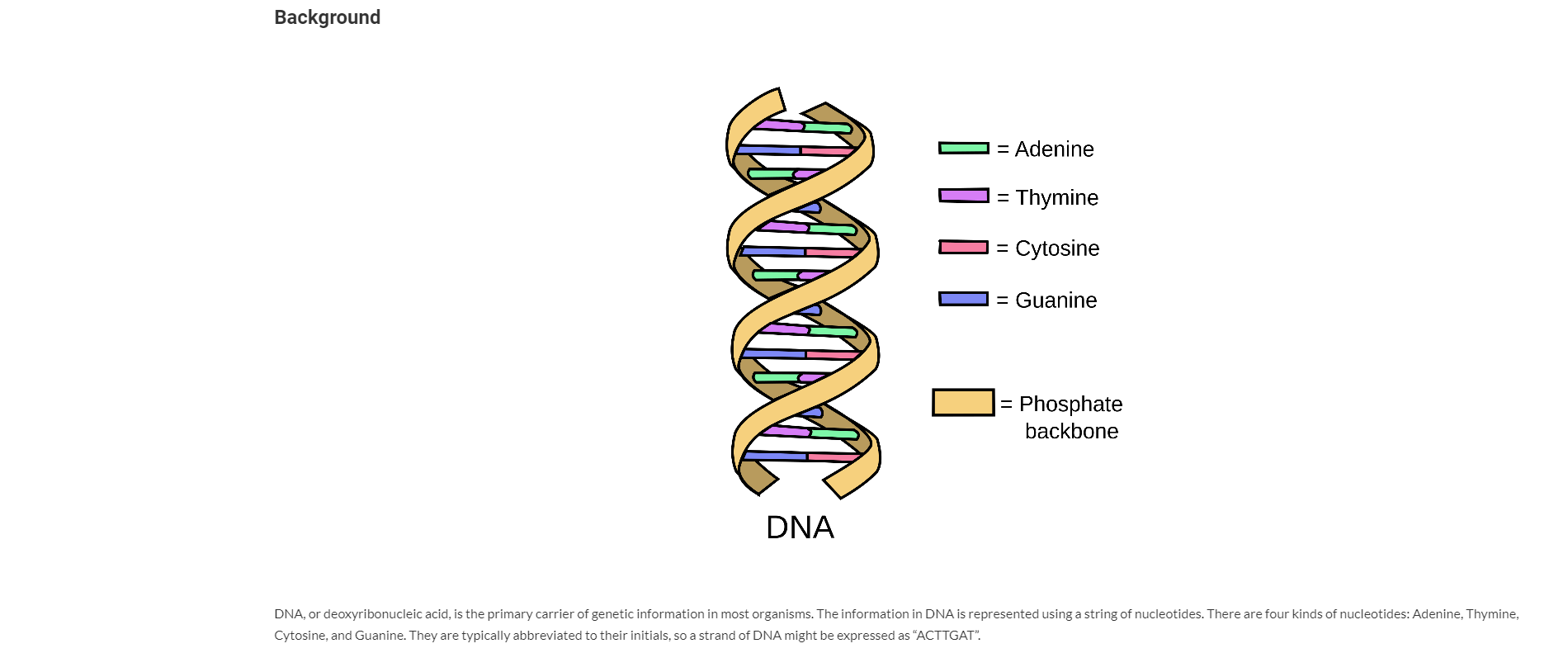
TEST_CASE( "Basic Constructor, getLength" ) {cout//This does NOT test to see if constructor stores the correct chars, just if it sets lengthDNAStrand str1("ACGTAGCT");REQUIRE( str1.getLength() == 8 );DNAStrand str2("AACC");REQUIRE( str2.getLength() == 4 );}TEST_CASE( "Basic Constructor, ==" ) {cout//Verify constructor actually stores the correct dataDNAStrand str1("ACGTAGCT");//If you put the == inside the REQUIRE it will not compile withoutbool isMatch = (str1 == DNAStrand("ACGTAGCT"));REQUIRE( isMatch );DNAStrand str2("AACC");bool isMatch2 = (str2 == DNAStrand("AACC"));bool isMatch3 = (str2 == DNAStrand("AAGC"));REQUIRE( isMatch2 );REQUIRE( !isMatch3 );}TEST_CASE( "==" ) {coutDNAStrand str("AACC");//Make sure unequal length stands don't cause == issuesbool isMatch1 = (str == DNAStrand("AAC"));bool isMatch2 = (str == DNAStrand("AACCT"));REQUIRE( !isMatch1 );REQUIRE( !isMatch2 );}TEST_CASE( "Copy constructor" ) {coutDNAStrand str1("ACGTAGCT");{/ew scopeDNAStrand str2(str1); //Call copy ctorbool isMatch = (str1 == str2);REQUIRE( isMatch );//str2 goes away when we leave this block...}bool isMatch2 = (str1 == DNAStrand("ACGTAGCT"));REQUIRE( isMatch2 );}TEST_CASE( "Assignment Operator" ) {coutDNAStrand str1("ACGTAGCT");{/ew scopeDNAStrand str2("AA");str2 = str1; //Use assignment opbool isMatch = (str2 == DNAStrand("ACGTAGCT"));REQUIRE( isMatch );//str2 goes away when we leave this block...}//Make sure str1 was not changedbool isMatch2 = (str1 == DNAStrand("ACGTAGCT"));REQUIRE( isMatch2 );//Verify self-assignment does no harm:{str1 = str1;}bool isMatch3 = (str1 == DNAStrand("ACGTAGCT"));REQUIRE( isMatch3 );}TEST_CASE( "operator[]" ) {coutDNAStrand str1("ACGTACGTACGT");REQUIRE( str1[0] == 'A' );REQUIRE( str1[5] == 'C' );DNAStrand str2("TTTT");REQUIRE( str2[0] == 'T' );REQUIRE( str2[2] == 'T' );//Check that excpetion is thrown if bad index providedint exceptionsCaught = 0;try {char c = str2[-1];} catch (out_of_range e) {exceptionsCaught++;}try {char c = str2[4];} catch (out_of_range e) {exceptionsCaught++;}REQUIRE( exceptionsCaught == 2 );}TEST_CASE( "operator+" ) {coutDNAStrand str1("AATT");DNAStrand str2("CCGG");DNAStrand str3 = str1 + str2;bool isMatch = (str3 == DNAStrand("AATTCCGG"));REQUIRE( isMatch );//Check originals aren't brokenbool isMatch2 = (str1 == DNAStrand("AATT"));bool isMatch3 = (str2 == DNAStrand("CCGG"));REQUIRE( isMatch2 );REQUIRE( isMatch3 );}TEST_CASE( "getComplement" ) {coutDNAStrand str1("ACTGG");DNAStrand str2 = str1.getComplement();bool isMatch1 = (str2 == DNAStrand("TGACC"));REQUIRE( isMatch1 );DNAStrand str3 = str2.getComplement(); //should be back where we startedbool isMatch2 = (str3 == DNAStrand("ACTGG"));REQUIRE( isMatch2 );}TEST_CASE( "OperatorcoutDNAStrand str1("AAC");DNAStrand str2("GT");stringstream result;resultREQUIRE( result.str() == "AAC GT" );}TEST_CASE( "substr" ) {coutDNAStrand str1("AACCGGTT");DNAStrand str2 = str1.substr(0,1);bool isMatch2 = (str2 == DNAStrand("A"));REQUIRE( isMatch2 );DNAStrand str3 = str1.substr(0,3);bool isMatch3 = (str3 == DNAStrand("AAC"));REQUIRE( isMatch3 );DNAStrand str4 = str1.substr(2,4);bool isMatch4 = (str4 == DNAStrand("CCGG"));REQUIRE( isMatch4 );DNAStrand str5 = str1.substr(2,6);bool isMatch5 = (str5 == DNAStrand("CCGGTT"));REQUIRE( isMatch5 );int exceptionsCaught = 0;try {//Check bad startstr1.substr(-1,4);} catch (out_of_range e) {exceptionsCaught++;}try {//Check bad lengthstr1.substr(3, -1);} catch (out_of_range e) {exceptionsCaught++;}try {//Check start + length invalidstr1.substr(3, 6);} catch (out_of_range e) {exceptionsCaught++;}REQUIRE( exceptionsCaught == 3 );}#ifndef DNASTRAND_H#define DNASTRAND_H#ifndef DNAStrand_H#define DNAStrand_H#includeusing namespace std;class DNAStrand {public:/*** @brief DNAStrand Builds a DNAStrand using the given C-String* @param startingString A null terminated char array** Throws a logic_error exception if the startingString contains anything but A, C, G, or T*/DNAStrand(const char* startingString);/*** @brief DNAStrand Copy Ctor* @param other DNAStrand to copy*/DNAStrand(const DNAStrand& other);/*** @brief operator = Assignment Operator for DNAStrand* @param other DNAStrand to duplicate* @return Reference to the string that was copied into*/DNAStrand& operator=(const DNAStrand& other);/*** @brief Destructor*/~DNAStrand();/*** @brief operator == Check if two DNAStrands have same contents* @param other DNAStrand to compate to* @return true if identical, false if not*/bool operator==(const DNAStrand& other) const;/*** @brief operator [] Access individual nucleotide from DNAStrand* @param index location to access* @return char value at index. Not returned by referece - no write access** Throws an out_of_range exception if the index is less than 0 or too large*/char operator[](int index) const;/*** @brief operator + Combine two DNAStrands* @param other DNAStrand to merge with this* @return DNAStrand with contents of current DNAStrand followed by contents of other*/DNAStrand operator+(const DNAStrand& other) const;//Declare as friend... declared after classfriend std::ostream& operator/*** @brief getLength gets length of string* @return length*/int getLength() const;/*** @brief getComplement gets Complement of the DNAStrand* @return DNAStrand that is complement of this** The complement is the DNAStrand that pairs with this one to make a double strand.* Any A's in originals must become T's in complement and vice verse* C's become G's and vice verse.** Original: ACCTG* Complement: TGGAC*/DNAStrand getComplement() const;/*** @brief substr Gets a DNAStrand that contains the requested portion of a DNAStrand* @param start index of first base to grab* @param length number of bases to include* @return DNAstrand** Throws an out_of_range exception if the index or length are less than 0* or start + length end up past the end of the array*/DNAStrand substr(int start, int length) const;private:/*** @brief DNAStrand Helper ctor - use in other functions to make a DNAStrand of given size that* then can be modified* @param length Size DNA String to create*/DNAStrand(int length);char* bases = nullptr; ///Tracks array contining a char arrayint length = 0; ///Length of the char array and DNAStrand};/*** @brief operator* @param dest stream to output to* @param source DNAStrand to output* @return dest stream returned by reference for chaining of*/std::ostream& operator#endif#endif // DNASTRAND_HBackground = Adenine - Thymine = - Cytosine ININING = Guanine = Phosphate backbone DNA DNA, or deoxyribonucleic acid, is the primary carrier of genetic information in most organisms. The information in DNA is represented using a string of nucleotides. There are four kinds of nucleotides: Adenine, Thymine, Cytosine, and Guanine. They are typically abbreviated to their initials, so a strand of DNA might be expressed as "ACTTGAT". These strands join together with a complementary strand to form DNA's famous double helix shape. Adenine and Thymine always pair up and Cytosine and Guanine always pair up. Thus, the strand: ACTTGAT Would always have a complementary strand: TGAACTA Where A in the original lines up with Tin the new strand, C in the original matches G in the new strand, etc... Assignment Instructions Submit files: DNAStrand.cpp, DNAStrandTester.cpp I should be able to add my own copies of DNAStrand.h and doctest.h and build your project in the Chemeketa Development Environment with the following command: g++ -g -std=C++11 DNAStrand.cpp DNAStrandTester.cpp -o program.exe Your task is to make a class that represent a DNA strand. A DNAStrand will track its length and a character array to store the bases (which will be stored using the characters A, C, G, and T). We will normally create a DNAStrand by doing something like: DNAStrand strand1("ACTGAGATA"); DNAStrand will also provide various operators and functions for doing things like combining two strands, getting the complement of a strand or searching for one strand inside another. You are provided a DNAStrand.h and DNAStrand Tester.cpp. You should not modify DNAStrand.h-you will not be turning it in; I will supply my own original copy. You should only modify DNAStrandTester.cpp to comment out entire TEST_CASEs that do not compile with your code or that cause a crash. Do not modify a TEST_CASE in any way except to comment the entire thing out. Leave in any TEST CASES that compile and run, even if they fail. Passing Test > Failing Test > Commented-Out Test > Does not Compile See below for tips on specific functions. Getting Started You should make a Unit Test project and add the two given files to it. (You do not need a "normal" project; you can do everything in the unit test project.) You will need to make your own DNAStrand.cpp file. Comment out all the tests but the first one in DNAStrandTester. Implement the constructor and getLength and work until you pass test 1. Then, work on one new test at a time, implementing only the new functions needed to pass that test. Memory Management You should use the Chemeketa Development Environment to test your code for leaks or other memory errors. If you place your project folder in the Vagrant shared folder, you can open it in QTCreator to work on it in your host system, and at the same time, access the code within the virtual machine to use DrMemory on it. Work on the code in QTCreator-you can build and run the project from there and it will be built to the debug/folder). Then periodically, switch to the virtual machine and build your program to a linux/folder and test that build inside the VM with DrMemory. Build (from the directory with your code-first make a folder there called linux): g++ -std=C++11 DNAStrand.cpp DNAStrandTester.cpp -o linux/program.exe Test (from that same directory): drmemory -- linux/program.exe DrMemory should give your program a clean bill of health. Any errors reported by DrMemory will result in a significant deduction, even if the program appears to work fine despite of them. In particular, watch out for memory leaks. Memory leaks will not cause any visible errors in your program-the only way to know that you have a leak is to use DrMemory (or run your code long enough that you can measure a steady increase in used memory). Remember, DrMemory shows you where it detected an issue. Often, the place where you caused the issue is somewhere else! For example, maybe your constructor did not allocate an array but that was not a problem until later when you went to write to that array. Function Tips Design Note: Most of the functions we will write are non-modifying. Once we make a DNAStrand, there is no way to change it other than the assignment operator. If we want to do something like add two strands, the originals will remain unmodified and we will end up with a new strand that represents them stuck together. Objects that can't be changed are known as immutable. Immutable objects are safer to pass around with pointers or references (since they are essentially always const). They also make it easier to write parallel code (no worries that one thread will be working with an object while another modifies it). The downside of immutable objects is we end up making lots of copies since every "changereally just makes a new object. If we wanted the class to be completely immutable, we would not provide an assignment operator. However, remember to allocate the "new" objects on the stack as local variables and return them. Do not use the new keyword, or the objects would be allocated on the heap and you would have to pass them around as pointers and remember to explicitly delete them. General restriction: you may not use std::string in this project. The point of this assignment is to learn about what is involved in making a string-like object and doing your own memory management. DNAStrand (const char* startingString) Our basic constructor-it takes a char array (C-String) and sets the length and creates an array that holds the bases listed in the C-String. You can use strlen fromto figure out the length of the parameter, or write a loop that looks for the char. (See week 8 of 161 for cstring material). You can use strlen from to figure out the length of the parameter, or write a loop that looks for the lo char. (See week 8 of 161 for cstring material). Your DNAStrand's char array does not have to end in a null character. That means you should NOT try to use cstring functions on the array you are storing-they expect char *s to be marked with a null char where the valid data ends. int getLength() const Just return length. bool operator==(const DNAStrand& other) const Checks to see if the strands contain an identical list of bases. (i.e. "ATTAG" and "ATTAG"). Needed by the first test and most others). You can't just compare the arrays themselves with == that will just see if the two DNAStrands are pointing to the same memory. You have to compare the individual items in the arrays. What is an easy way to tell that two strands can't possibly be identical? ~DNAStrand() Destructor. Not tested directly-but every test will leak memory without this. Verify it works by running your program with drmemory. DNAStrand(const DNAStrand& other) and DNAStrand& operator=(const DNAStrand& other) Copy constructor and assignment operator. Tested in two unit tests. These are another big area for memory issues-test with DrMemory! char operator[](int index) const Gets a particular base. Return type is char instead of char& so that we can only read the bases and can't modify them. (Using something like strand[2] = 'T' won't work unless operator[] returns a reference.) This needs to throw an out_of_range exception if given a bad index. DNAStrand(int length) This private constructor builds a DNAStrand with the given length and should allocate an array big enough to hold that many chars (and not initialize them or init to all O). Having this constructor will make other functions easier to write as they will be able to do something like: DNAStrand temp(20); // I know it should be 2e long //use loop to set temp's bases return tempi It is private as we don't want other code using this special helper constructor. There are no tests for this (since it is private). But you will probably want to use it as part of any function that returns a DNAStrand. Chemeketa CS People Course Resources CS Advising DNAStrand (const DNAStrand& other) and DNAStrand& operator=(const DNAStrand& other) Copy constructor and assignment operator. Tested in two unit tests. These are another big area for memory issues-test with DrMemory! char operator[](int index) const Gets a particular base. Return type is char instead of char& so that we can only read the bases and can't modify them. (Using something like strand[2] = 'T' won't work unless operator[] returns a reference.) This needs to throw an out_of_range exception if given a bad index. DNAStrand(int length) This private constructor builds a DNAStrand with the given length and should allocate an array big enough to hold that many chars (and not initialize them or init to all O). Having this constructor will make other functions easier to write as they will be able to do something like: DNAStrand temp(20); // I know it should be 20 long //use loop to set temp's bases return temp; It is private as we don't want other code using this special helper constructor. There are no tests for this (since it is private). But you will probably want to use it as part of any function that returns a DNAStrand. operator+ and getComplement Check the.h for documentation. Note that all of these return a new object that has the desired DNAStrand. Don't try to modify the current strand. Each has their own test. You will probably need/want the DNAString(int length) constructor as a helper for these. operator Failing Test > Commented-Out Test > Does not Compile See below for tips on specific functions. Getting Started You should make a Unit Test project and add the two given files to it. (You do not need a "normal" project; you can do everything in the unit test project.) You will need to make your own DNAStrand.cpp file. Comment out all the tests but the first one in DNAStrandTester. Implement the constructor and getLength and work until you pass test 1. Then, work on one new test at a time, implementing only the new functions needed to pass that test. Memory Management You should use the Chemeketa Development Environment to test your code for leaks or other memory errors. If you place your project folder in the Vagrant shared folder, you can open it in QTCreator to work on it in your host system, and at the same time, access the code within the virtual machine to use DrMemory on it. Work on the code in QTCreator-you can build and run the project from there and it will be built to the debug/folder). Then periodically, switch to the virtual machine and build your program to a linux/folder and test that build inside the VM with DrMemory. Build (from the directory with your code-first make a folder there called linux): g++ -std=C++11 DNAStrand.cpp DNAStrandTester.cpp -o linux/program.exe Test (from that same directory): drmemory -- linux/program.exe DrMemory should give your program a clean bill of health. Any errors reported by DrMemory will result in a significant deduction, even if the program appears to work fine despite of them. In particular, watch out for memory leaks. Memory leaks will not cause any visible errors in your program-the only way to know that you have a leak is to use DrMemory (or run your code long enough that you can measure a steady increase in used memory). Remember, DrMemory shows you where it detected an issue. Often, the place where you caused the issue is somewhere else! For example, maybe your constructor did not allocate an array but that was not a problem until later when you went to write to that array. Function Tips Design Note: Most of the functions we will write are non-modifying. Once we make a DNAStrand, there is no way to change it other than the assignment operator. If we want to do something like add two strands, the originals will remain unmodified and we will end up with a new strand that represents them stuck together. Objects that can't be changed are known as immutable. Immutable objects are safer to pass around with pointers or references (since they are essentially always const). They also make it easier to write parallel code (no worries that one thread will be working with an object while another modifies it). The downside of immutable objects is we end up making lots of copies since every "changereally just makes a new object. If we wanted the class to be completely immutable, we would not provide an assignment operator. However, remember to allocate the "new" objects on the stack as local variables and return them. Do not use the new keyword, or the objects would be allocated on the heap and you would have to pass them around as pointers and remember to explicitly delete them. General restriction: you may not use std::string in this project. The point of this assignment is to learn about what is involved in making a string-like object and doing your own memory management. DNAStrand (const char* startingString) Our basic constructor-it takes a char array (C-String) and sets the length and creates an array that holds the bases listed in the C-String. You can use strlen from to figure out the length of the parameter, or write a loop that looks for the char. (See week 8 of 161 for cstring material). You can use strlen from to figure out the length of the parameter, or write a loop that looks for the lo char. (See week 8 of 161 for cstring material). Your DNAStrand's char array does not have to end in a null character. That means you should NOT try to use cstring functions on the array you are storing-they expect char *s to be marked with a null char where the valid data ends. int getLength() const Just return length. bool operator==(const DNAStrand& other) const Checks to see if the strands contain an identical list of bases. (i.e. "ATTAG" and "ATTAG"). Needed by the first test and most others). You can't just compare the arrays themselves with == that will just see if the two DNAStrands are pointing to the same memory. You have to compare the individual items in the arrays. What is an easy way to tell that two strands can't possibly be identical? ~DNAStrand() Destructor. Not tested directly-but every test will leak memory without this. Verify it works by running your program with drmemory. DNAStrand(const DNAStrand& other) and DNAStrand& operator=(const DNAStrand& other) Copy constructor and assignment operator. Tested in two unit tests. These are another big area for memory issues-test with DrMemory! char operator[](int index) const Gets a particular base. Return type is char instead of char& so that we can only read the bases and can't modify them. (Using something like strand[2] = 'T' won't work unless operator[] returns a reference.) This needs to throw an out_of_range exception if given a bad index. DNAStrand(int length) This private constructor builds a DNAStrand with the given length and should allocate an array big enough to hold that many chars (and not initialize them or init to all O). Having this constructor will make other functions easier to write as they will be able to do something like: DNAStrand temp(20); // I know it should be 2e long //use loop to set temp's bases return tempi It is private as we don't want other code using this special helper constructor. There are no tests for this (since it is private). But you will probably want to use it as part of any function that returns a DNAStrand. Chemeketa CS People Course Resources CS Advising DNAStrand (const DNAStrand& other) and DNAStrand& operator=(const DNAStrand& other) Copy constructor and assignment operator. Tested in two unit tests. These are another big area for memory issues-test with DrMemory! char operator[](int index) const Gets a particular base. Return type is char instead of char& so that we can only read the bases and can't modify them. (Using something like strand[2] = 'T' won't work unless operator[] returns a reference.) This needs to throw an out_of_range exception if given a bad index. DNAStrand(int length) This private constructor builds a DNAStrand with the given length and should allocate an array big enough to hold that many chars (and not initialize them or init to all O). Having this constructor will make other functions easier to write as they will be able to do something like: DNAStrand temp(20); // I know it should be 20 long //use loop to set temp's bases return temp; It is private as we don't want other code using this special helper constructor. There are no tests for this (since it is private). But you will probably want to use it as part of any function that returns a DNAStrand. operator+ and getComplement Check the.h for documentation. Note that all of these return a new object that has the desired DNAStrand. Don't try to modify the current strand. Each has their own test. You will probably need/want the DNAString(int length) constructor as a helper for these. operator
Step by Step Solution
There are 3 Steps involved in it
1 Expert Approved Answer
Step: 1 Unlock


Question Has Been Solved by an Expert!
Get step-by-step solutions from verified subject matter experts
Step: 2 Unlock
Step: 3 Unlock