Question: I need a working python script for forensic science . It should have a reading frame and should locate the amino acid 'M', in any
I need a working python script for forensic science . It should have a reading frame and should locate the amino acid 'M', in any DNA sequence and prints whatever is after that 'M' till it reaches a stop codon '-'. It should ask me input reading frame. please use the same terms as I used in the attached script below. I should be also to get ANY amino acids from any DNA sequence.
EXAMPLE 1, if the input is ''RLDMVGQ---KLVV'', the protein_finder function will return ['MVGQ']. Not an two empty strings '',''
2: input is 'KMQKKML' and the out put will be ['MQKKML']. I want the function to only read the first M, and treat every other M as normal and add to the output as normal M.
if I input empty string ' ', then the output should be [ ]. Not an error. if I input a reading frame without the 'M' amino acid (aa), then the out should be empty [ ].
I have added a sample starting function that u can modify. please use so that the script can read this dan function "ATGAAATGGCGAGGAAAGAAAGGGGCCTCGCTAGCAGCTGAGAA" and give me a list of 2 proteins in it.
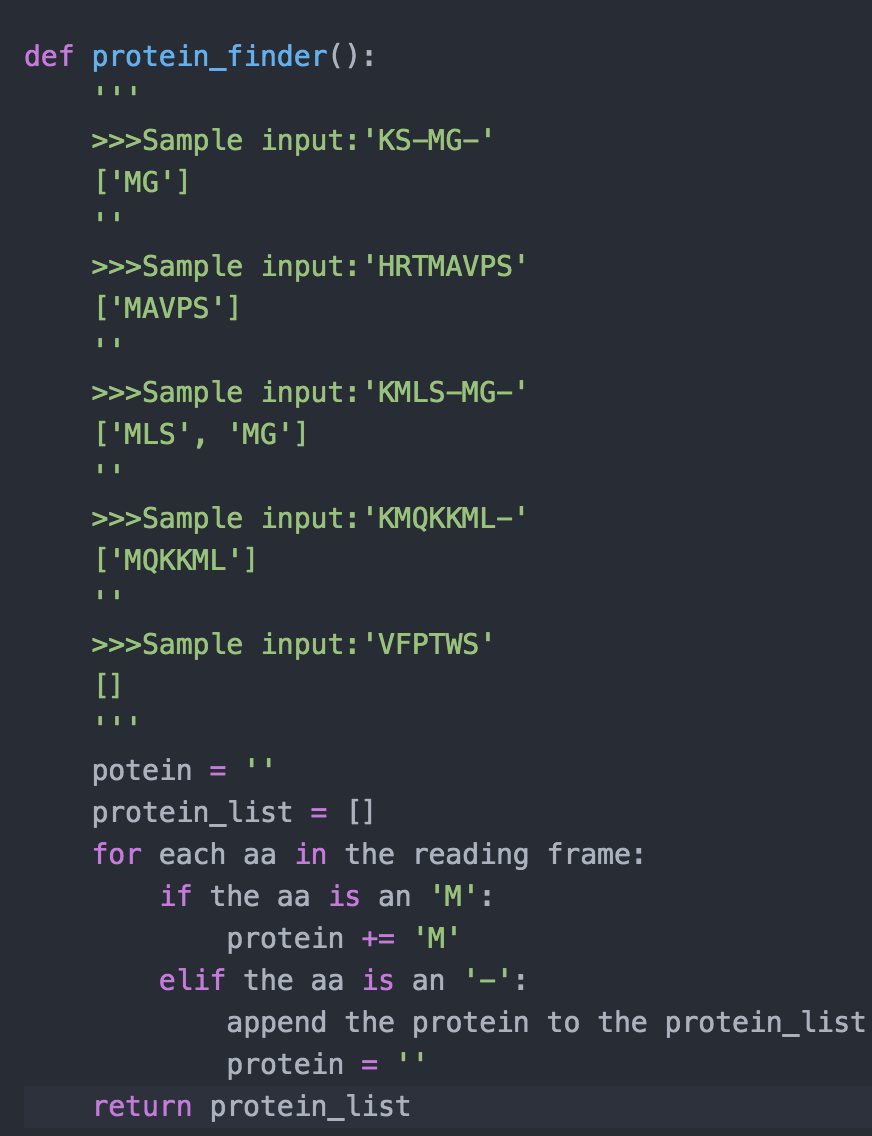
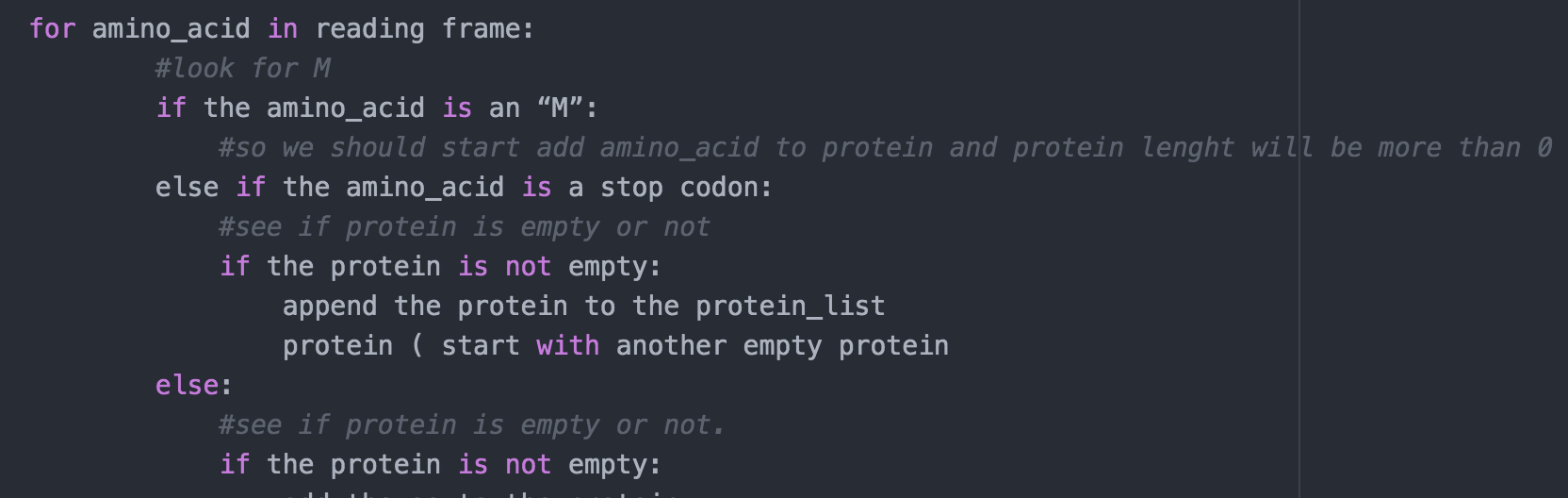
IN THIS SCRPIT position = reading_frame.find('M') IS NOT CORRECT. this will not be helpful. 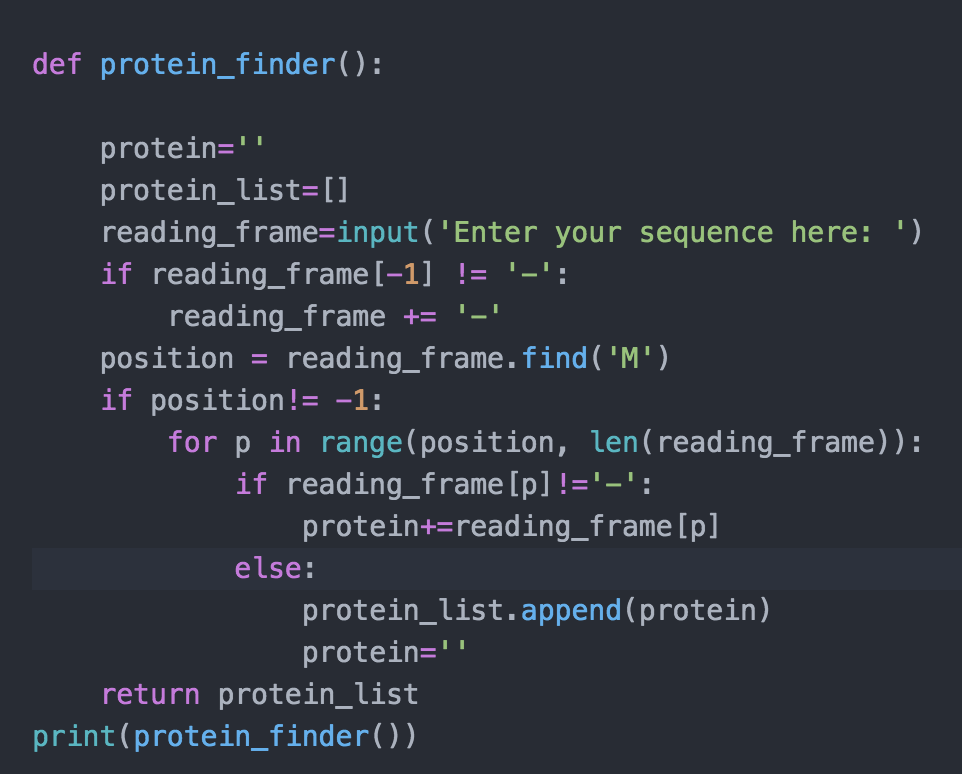
def protein_finder(): >>>Sample input: 'KS-MG-' ['MG'] >>>Sample input: 'HRTMAVPS' ['MAVPS'] >>>Sample input: 'KMLS-MG-' ['MLS', 'MG'] >>>Sample input: 'KMQKKML-' ['MQKKML'] >>>Sample input: 'VFPTWS' [] potein protein_list = [] for each aa in the reading frame: if the aa is an 'M': protein += 'M' elif the aa is an '-': append the protein to the protein_list protein return protein_list for amino acid in reading frame: #look for M if the amino_acid is an "M": #so we should start add amino acid to protein and protein lenght will be more than 0 else if the amino acid is a stop codon: #see if protein is empty or not if the protein is not empty: append the protein to the protein_list protein ( start with another empty protein else: #see if protein is empty or not. if the protein is not empty: def protein_finder(): protein='I protein_list=[] reading_frame=input('Enter your sequence here: ') if reading_frame[-1] != '-': reading_frame += position = reading_frame.find('M') if position!= -1: for p in range(position, len(reading_frame)): if reading_frame [p] !='-': protein+=reading_frame[p] else: protein_list.append(protein) protein=" return protein_list print(protein_finder())
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


