Question: instruction up top The Homework title is Implement class for manipulating DNA sequence in plain format I'm stuck on this part of the coding question.
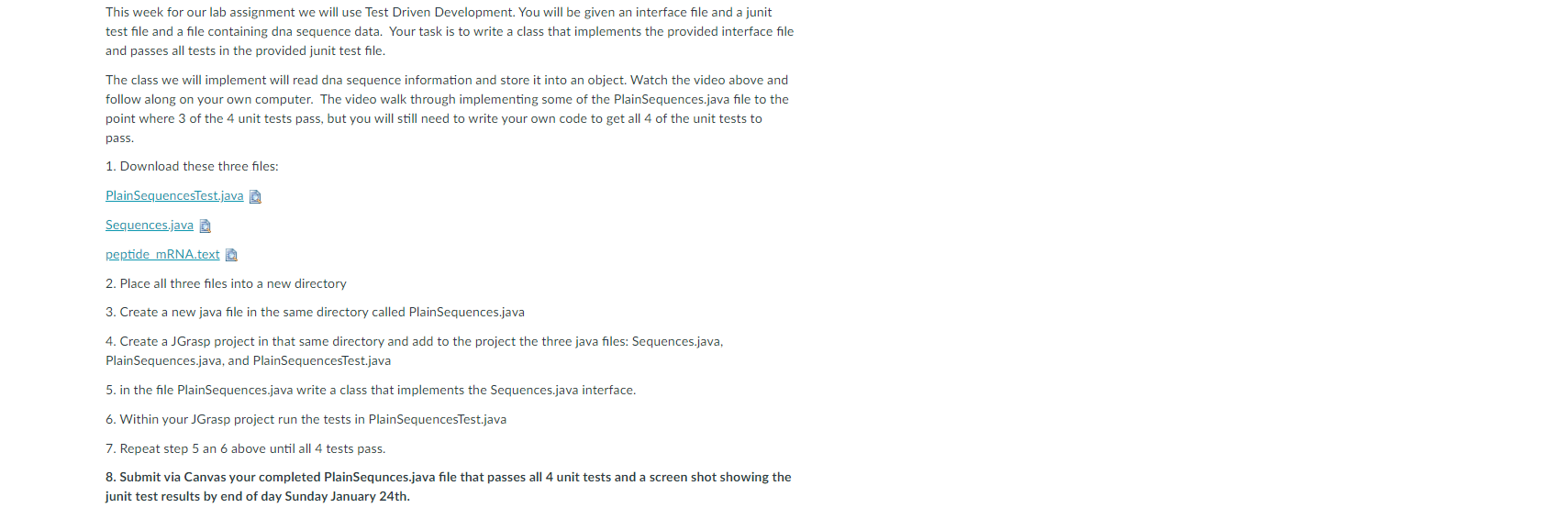
instruction up top
The Homework title is "Implement class for manipulating DNA sequence in plain format"
I'm stuck on this part of the coding question.
The first picture is where I should be writing the codes for the test code in picture 2
(It would be helpful if you explain your answer, since my teacher did not teach at all.)
Thank you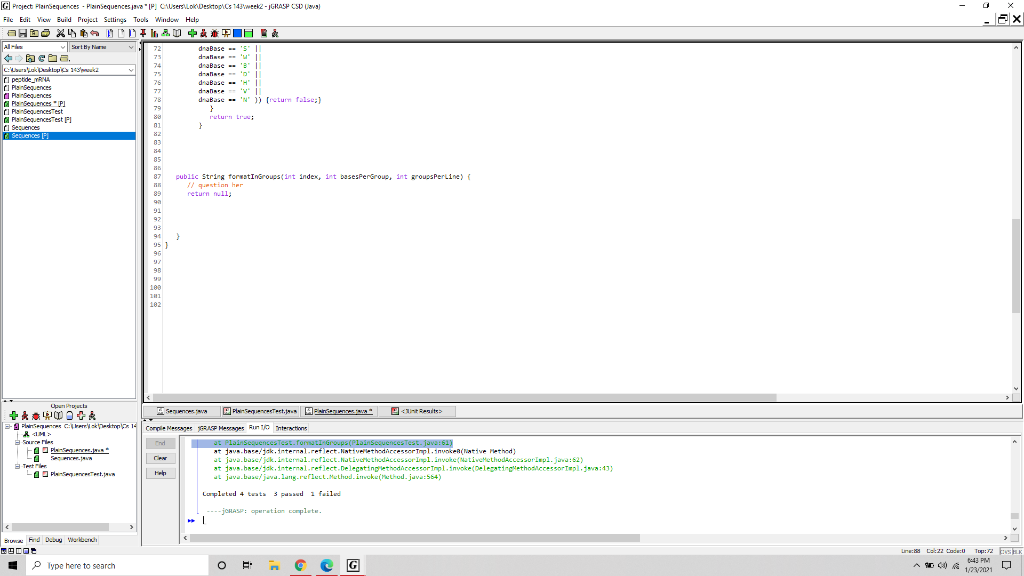
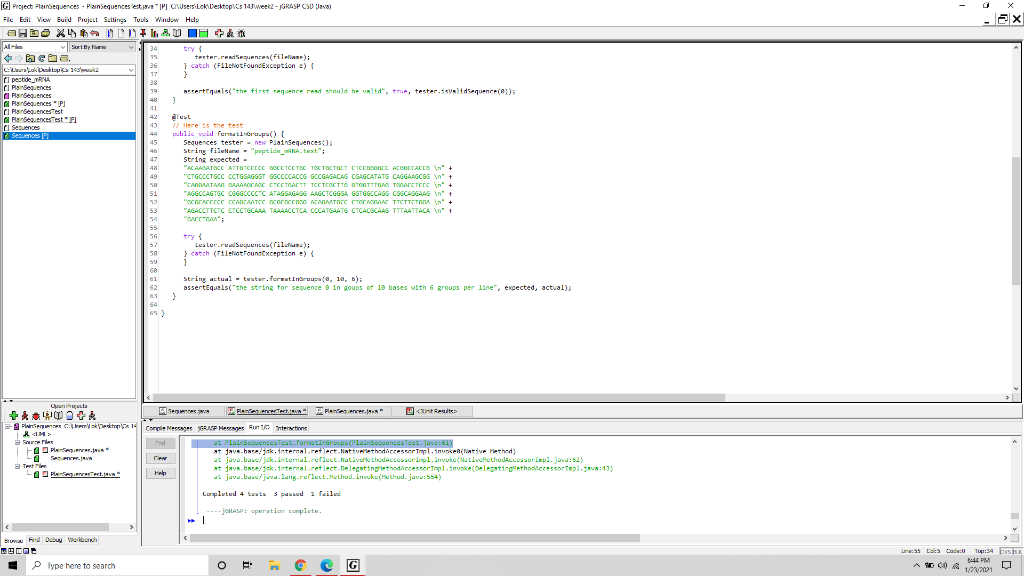
This week for our lab assignment we will use Test Driven Development. You will be given an interface file and a junit test file and a file containing dna sequence data. Your task is to write a class that implements the provided interface file and passes all tests in the provided junit test file. The class we will implement will read dna sequence information and store it into an object. Watch the video above and follow along on your own computer. The video walk through implementing some of the PlainSequences.java file to the point where 3 of the 4 unit tests pass, but you will still need to write your own code to get all 4 of the unit tests to pass. 1. Download these three files: Plain SequencesTest.java Sequences.java peptide mRNA.text 2 2. Place all three files into a new directory 3. Create a new java file in the same directory called PlainSequences.java 4. Create a Grasp project in that same directory and add to the project the three java files: Sequences.java, PlainSequences.java, and PlainSequencesTest.java 5. in the file PlainSequences.java write a class that implements the sequences.java interface. 6. Within your Grasp project run the tests in PlainSequencesTest.java 7. Repeat step 5 an 6 above until all 4 tests pass. 8. Submit via Canvas your completed PlainSequnces.java file that passes all 4 unit tests and a screen shot showing the junit test results by end of day Sunday January 24th. droBase - G Project Plainsequences - Plainsequences". Users LDK Desktop CS 143'week2-GRASP OSD Vaval File Edit View Buid Project Setting Tools Window Hdp A XUDDI 204&TIH R& al Sart Hy 72 51 75 dance -- 24 daBase churakka 75 daar- Il peptide YANA 29 de Base - H i Plainsequence 77 delete Planequences 28 Paneg.ences d-Base - 'N' ) [return false) 79 } Pequences est rului ! PisinSequencesesti 01 } Secuenos 85 87 public String forwatInGroups(int index, int besesperGroup, int groupsPerLine) 11 stinn her return null; ga 91 92 95 90 951 > 180 182 Openisula Preces. We CAVA Untegits 3- PR Credit Piedi 14 TAM Sarees PAPY. SPAVA BT - Pleqerecer Comple essages RISP Merges Run 10 Interactions a secuences Test.mot Groups (Plain SecuencesTest.java:61) at java.haresk.internal.reflect.NativelethodAccescortpl.invoke(Native Method) at java.base/jak.internal.reflect.NativethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:52) at java.back 1ternal.reflect.Delegating lethodAccescortpl.invoke(DelegatingMethosilccessoriepl.wa:43) Help at java.us/jw.lang.reflect.Method.invoke(Method.java:554) CAN Lumplutud + tuats Spa i fall ----RASP: puration completu. Bw Frid Dulu Workbench LUDOS Type here to search Line Lot 22 Gade ES 9 G ACO 1443 PM 1/23/201 Sarthy 36 17 19 48 43 G Project Plainsequences - Plainsequences est. "IP CUsers' Lok Desktops 145 Wee2 - GRASP CSD Pava) File Edit View Build Project Settings Tools Window Help A XUDDI 2.0 IH + al terter.redes(fileliner); tester cultural 1 catch (FileNotFoundException e) } Il pertide 38 i Planequences Planeering accert quals("the first sequence red should be valid", trie, tester.altd sequence()); Pequences 2 1 41 i nequences est ATO Ploegerxes est Secuenos 1 Here is the test public void urna trup() { Sequences tester - new Plainsequences); String film "peptide_text": String expected "ACARCATOCC ATTITCODE BOCETCETOC TOC TIC TOET ETCCECODEC ACCOCCADA I 49 "CTGCCCTSCC CCTGSAGGST GOCCOCACOG GCCGAGACAG CSAGCATATG CAGGAAGCGS + "CANTATARS CAROCACC CTCCTCAETT TCCTGOETTG ETOOTITIAS TECACTCECI + 51 "AGGCCAGTGC CGGGOCCCTC ATAGSAGASG ALGCTCGGSA GSTGGOCASS CSSCASGAAG " + 12 "ecoracce CACAATCC crecen ACARAATIC CTOCAGAN TTCTTCTORA In" + 53 AGACCTTCTC CTCCTGCASA TAAAACCTCA CCCATGAATG CTCACSCALS TTTAATTACA" + 54 55 SG 52 Lesler.read() 50 } catch (FileliotFoundException e) { ] GB String actual - tester, forsetinn, 18, ); assertEquals("the string for sequence e in goups of 18 bases with 6 groups per line", expected, actual); } 45 46 Upunta Plans verehe Planes ." Resit 3- PR Credit Comple Messages RASP Meg in 10 interactions A SIM at Pla Sequences Test Tort Groups (Pla SecuencesTest.java:61) - Pia. at java.hacers.internal.reflect.Native lethedilccescortpl.invoke(Native Method) @ec .. CAN at java.base/jak.internal.reflect.NativcethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:52) at java.bacetek, internal.reflect.Delegating lethodccessorImpl.invoke(Delesting ethosilccessoriepl.wa:43) Help at java.us/jww.lang.reflect.Method.invoke(Method.java:554) Lapletud + tuata pada lelle ----RASP: puration completu. BT Bw Frid Dulu Workbench LUDOS Type here to search Line Cats Case Top:34 E 0:44 PM 1/23/201 ES 9 G ASCO This week for our lab assignment we will use Test Driven Development. You will be given an interface file and a junit test file and a file containing dna sequence data. Your task is to write a class that implements the provided interface file and passes all tests in the provided junit test file. The class we will implement will read dna sequence information and store it into an object. Watch the video above and follow along on your own computer. The video walk through implementing some of the PlainSequences.java file to the point where 3 of the 4 unit tests pass, but you will still need to write your own code to get all 4 of the unit tests to pass. 1. Download these three files: Plain SequencesTest.java Sequences.java peptide mRNA.text 2 2. Place all three files into a new directory 3. Create a new java file in the same directory called PlainSequences.java 4. Create a Grasp project in that same directory and add to the project the three java files: Sequences.java, PlainSequences.java, and PlainSequencesTest.java 5. in the file PlainSequences.java write a class that implements the sequences.java interface. 6. Within your Grasp project run the tests in PlainSequencesTest.java 7. Repeat step 5 an 6 above until all 4 tests pass. 8. Submit via Canvas your completed PlainSequnces.java file that passes all 4 unit tests and a screen shot showing the junit test results by end of day Sunday January 24th. droBase - G Project Plainsequences - Plainsequences". Users LDK Desktop CS 143'week2-GRASP OSD Vaval File Edit View Buid Project Setting Tools Window Hdp A XUDDI 204&TIH R& al Sart Hy 72 51 75 dance -- 24 daBase churakka 75 daar- Il peptide YANA 29 de Base - H i Plainsequence 77 delete Planequences 28 Paneg.ences d-Base - 'N' ) [return false) 79 } Pequences est rului ! PisinSequencesesti 01 } Secuenos 85 87 public String forwatInGroups(int index, int besesperGroup, int groupsPerLine) 11 stinn her return null; ga 91 92 95 90 951 > 180 182 Openisula Preces. We CAVA Untegits 3- PR Credit Piedi 14 TAM Sarees PAPY. SPAVA BT - Pleqerecer Comple essages RISP Merges Run 10 Interactions a secuences Test.mot Groups (Plain SecuencesTest.java:61) at java.haresk.internal.reflect.NativelethodAccescortpl.invoke(Native Method) at java.base/jak.internal.reflect.NativethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:52) at java.back 1ternal.reflect.Delegating lethodAccescortpl.invoke(DelegatingMethosilccessoriepl.wa:43) Help at java.us/jw.lang.reflect.Method.invoke(Method.java:554) CAN Lumplutud + tuats Spa i fall ----RASP: puration completu. Bw Frid Dulu Workbench LUDOS Type here to search Line Lot 22 Gade ES 9 G ACO 1443 PM 1/23/201 Sarthy 36 17 19 48 43 G Project Plainsequences - Plainsequences est. "IP CUsers' Lok Desktops 145 Wee2 - GRASP CSD Pava) File Edit View Build Project Settings Tools Window Help A XUDDI 2.0 IH + al terter.redes(fileliner); tester cultural 1 catch (FileNotFoundException e) } Il pertide 38 i Planequences Planeering accert quals("the first sequence red should be valid", trie, tester.altd sequence()); Pequences 2 1 41 i nequences est ATO Ploegerxes est Secuenos 1 Here is the test public void urna trup() { Sequences tester - new Plainsequences); String film "peptide_text": String expected "ACARCATOCC ATTITCODE BOCETCETOC TOC TIC TOET ETCCECODEC ACCOCCADA I 49 "CTGCCCTSCC CCTGSAGGST GOCCOCACOG GCCGAGACAG CSAGCATATG CAGGAAGCGS + "CANTATARS CAROCACC CTCCTCAETT TCCTGOETTG ETOOTITIAS TECACTCECI + 51 "AGGCCAGTGC CGGGOCCCTC ATAGSAGASG ALGCTCGGSA GSTGGOCASS CSSCASGAAG " + 12 "ecoracce CACAATCC crecen ACARAATIC CTOCAGAN TTCTTCTORA In" + 53 AGACCTTCTC CTCCTGCASA TAAAACCTCA CCCATGAATG CTCACSCALS TTTAATTACA" + 54 55 SG 52 Lesler.read() 50 } catch (FileliotFoundException e) { ] GB String actual - tester, forsetinn, 18, ); assertEquals("the string for sequence e in goups of 18 bases with 6 groups per line", expected, actual); } 45 46 Upunta Plans verehe Planes ." Resit 3- PR Credit Comple Messages RASP Meg in 10 interactions A SIM at Pla Sequences Test Tort Groups (Pla SecuencesTest.java:61) - Pia. at java.hacers.internal.reflect.Native lethedilccescortpl.invoke(Native Method) @ec .. CAN at java.base/jak.internal.reflect.NativcethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:52) at java.bacetek, internal.reflect.Delegating lethodccessorImpl.invoke(Delesting ethosilccessoriepl.wa:43) Help at java.us/jww.lang.reflect.Method.invoke(Method.java:554) Lapletud + tuata pada lelle ----RASP: puration completu. BT Bw Frid Dulu Workbench LUDOS Type here to search Line Cats Case Top:34 E 0:44 PM 1/23/201 ES 9 G ASCO
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


