Question: Let's say you have nucleotide sequence BLAST database containing 10,000,000 sequences averaging 100 nucleotides in length. You are interested in BLASTing another sequence that is
Let's say you have nucleotide sequence BLAST database containing 10,000,000 sequences averaging 100 nucleotides in length. You are interested in BLASTing another sequence that is 20-nt in length. What E-value would you expect if the bit-score (S' in the notes) of the top hit is 5? What E-value would you expect if the bit-score (S' in the notes) of the top hit is 35? Hint: You can raise a number to a power in python with the double-asterisk operator. So 2-100 would be written in python as:
>>> 2**-100 7.888609052210118e-31
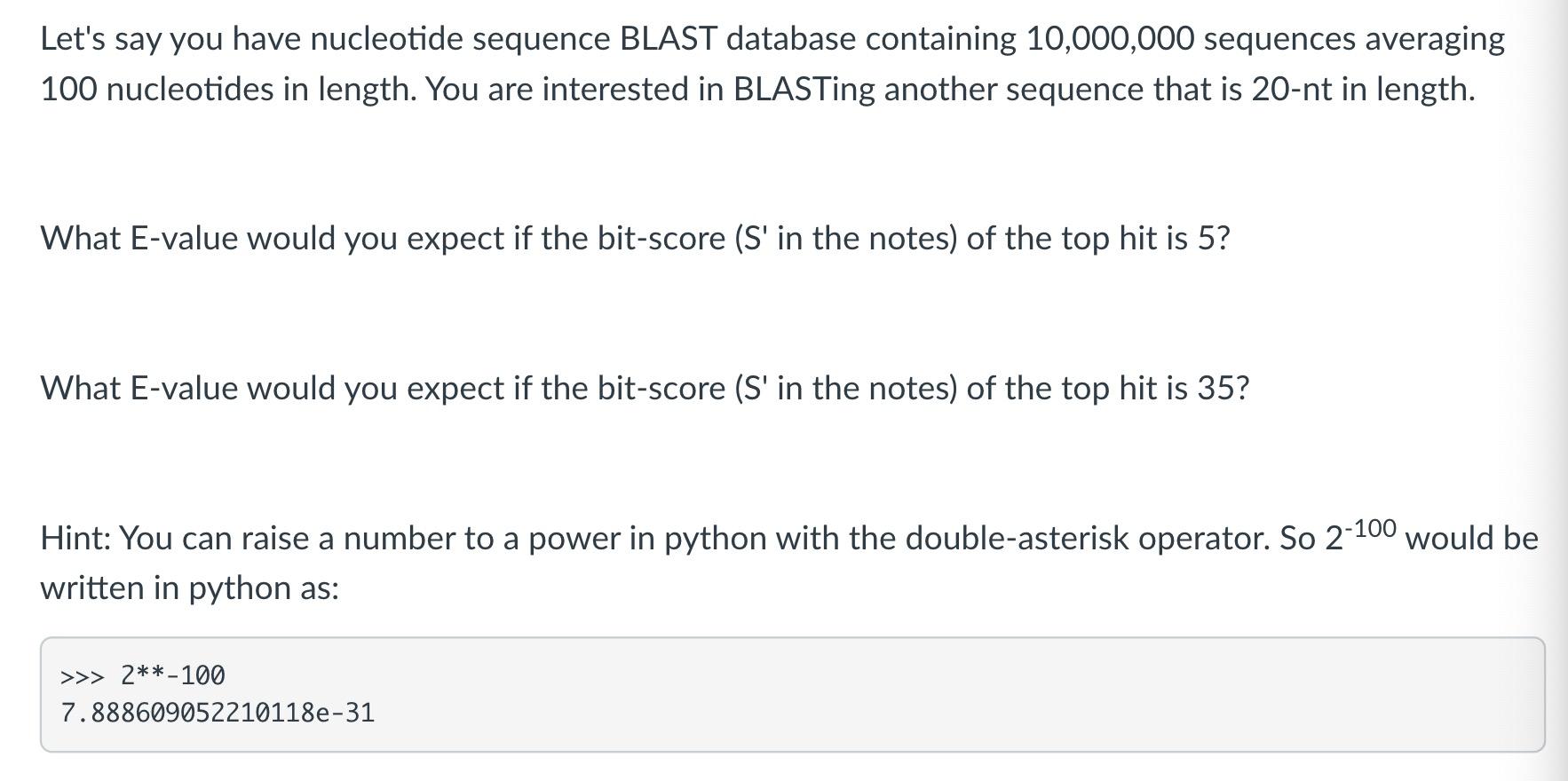
Let's say you have nucleotide sequence BLAST database containing 10,000,000 sequences averaging 100 nucleotides in length. You are interested in BLASTing another sequence that is 20-nt in length. What E-value would you expect if the bit-score (S' in the notes) of the top hit is 5? What E-value would you expect if the bit-score (S' in the notes) of the top hit is 35? Hint: You can raise a number to a power in python with the double-asterisk operator. So 2-100 would be written in python as: >>> 2**-100 7.888609052210118e-31 Let's say you have nucleotide sequence BLAST database containing 10,000,000 sequences averaging 100 nucleotides in length. You are interested in BLASTing another sequence that is 20-nt in length. What E-value would you expect if the bit-score (S' in the notes) of the top hit is 5? What E-value would you expect if the bit-score (S' in the notes) of the top hit is 35? Hint: You can raise a number to a power in python with the double-asterisk operator. So 2-100 would be written in python as: >>> 2**-100 7.888609052210118e-31
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


