Question: Python 3 def # Given a DNA sequence in string object, this function will not return anything, but it will # print out the 6
Python 3

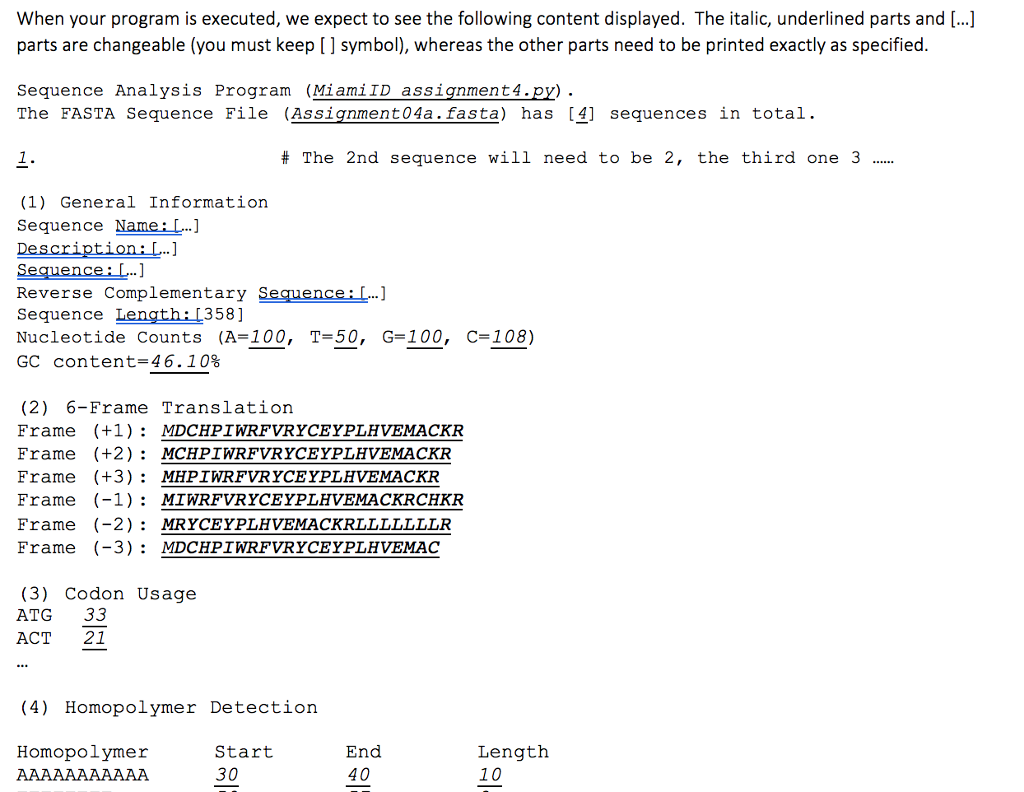
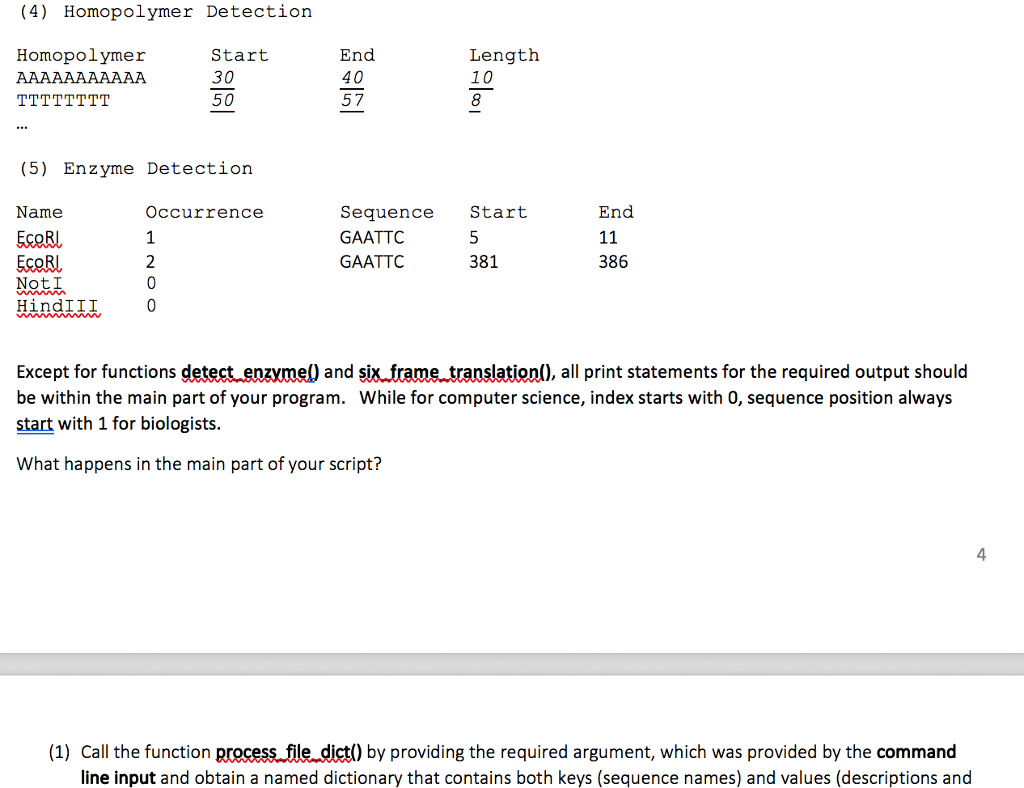
def # Given a DNA sequence in string object, this function will not return anything, but it will # print out the 6 frame translation results (eg, 6 proteins sequences). In this function # you can call translation() 6 times, obtain 6 protein sequences, and print them. Also, # you need to call cencomp..seal) to get the reverse complementary sequence for the # given DNA sequence. When your program is executed, we expect to see the following content displayed. The italic, underlined parts and [...] parts are changeable (you must keep [ ] symbol), whereas the other parts need to be printed exactly as specified Sequence Analysis Program (MiamiID assignment 4.py) The FASTA Seguence File (Assignment04a. fasta) has [41 sequences in total. # The 2nd sequence will need to be 2, the third one 3 (1) General Information Sequence Name... Reverse Complementary Sequence:L Sequence Length: 1358] Nucleotide Counts (A-100, T=50, GC content=46.10% ] G-100, C-108) (2) 6-Frame Translation Frame (+1MDCHPIWRFVRYCEYPLHVEMACKR Frame (+2): MCHPIWRFVRYCEYPLHVEMACKR Frame (+3)MHPIWRFVRYCEYPLHVEMACKR Frame (-1) MIWRFVRYCEYPLHVEMACKRCHKR Frame (-2) : MRYCEYPLHVEMACKRLLLLLLLR Frame (-3)MDCHPIWRFVRYCEYPLHVEMAC (3) Codon Usage ATG 33 ACT 21 (4) Homopolymer Detection Homopolymer Start 30 End 40 Length 10 (4) Homopolymer Detection Homopolymer Start 30 50 End 40 57 Length 10 8 (5) Enzyme Detection Name EcoRl EcoRl Occurrence End Seguence Start GAATTC GAATTC 381 386 NotI HindIII Except for functions detect enzymel) and six frame translation(), all print statements for the required output should be within the main part of your program. While for computer science, index starts with 0, sequence position always start with 1 for biologists r er ste nd s What happens in the main part of your script? 4 (1) Call the function process file dict() by providing the required argument, which was provided by the command line input and obtain a named dictionary that contains both keys (sequence names) and values (descriptions and def # Given a DNA sequence in string object, this function will not return anything, but it will # print out the 6 frame translation results (eg, 6 proteins sequences). In this function # you can call translation() 6 times, obtain 6 protein sequences, and print them. Also, # you need to call cencomp..seal) to get the reverse complementary sequence for the # given DNA sequence. When your program is executed, we expect to see the following content displayed. The italic, underlined parts and [...] parts are changeable (you must keep [ ] symbol), whereas the other parts need to be printed exactly as specified Sequence Analysis Program (MiamiID assignment 4.py) The FASTA Seguence File (Assignment04a. fasta) has [41 sequences in total. # The 2nd sequence will need to be 2, the third one 3 (1) General Information Sequence Name... Reverse Complementary Sequence:L Sequence Length: 1358] Nucleotide Counts (A-100, T=50, GC content=46.10% ] G-100, C-108) (2) 6-Frame Translation Frame (+1MDCHPIWRFVRYCEYPLHVEMACKR Frame (+2): MCHPIWRFVRYCEYPLHVEMACKR Frame (+3)MHPIWRFVRYCEYPLHVEMACKR Frame (-1) MIWRFVRYCEYPLHVEMACKRCHKR Frame (-2) : MRYCEYPLHVEMACKRLLLLLLLR Frame (-3)MDCHPIWRFVRYCEYPLHVEMAC (3) Codon Usage ATG 33 ACT 21 (4) Homopolymer Detection Homopolymer Start 30 End 40 Length 10 (4) Homopolymer Detection Homopolymer Start 30 50 End 40 57 Length 10 8 (5) Enzyme Detection Name EcoRl EcoRl Occurrence End Seguence Start GAATTC GAATTC 381 386 NotI HindIII Except for functions detect enzymel) and six frame translation(), all print statements for the required output should be within the main part of your program. While for computer science, index starts with 0, sequence position always start with 1 for biologists r er ste nd s What happens in the main part of your script? 4 (1) Call the function process file dict() by providing the required argument, which was provided by the command line input and obtain a named dictionary that contains both keys (sequence names) and values (descriptions and
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


