Question: The following corresponds to python programming. Will upvote, thx. 1) We are simplifying the create_mRNA() function. Specifically, we want to implement it in a single
The following corresponds to python programming. Will upvote, thx.
1) We are simplifying the create_mRNA() function. Specifically, we want to implement it in a single line. But there are errors. Find what went wrong and fix them.
a) PLEASE WRITE THE COMPLETED CODE BELOW IN YOUR ANSWER.
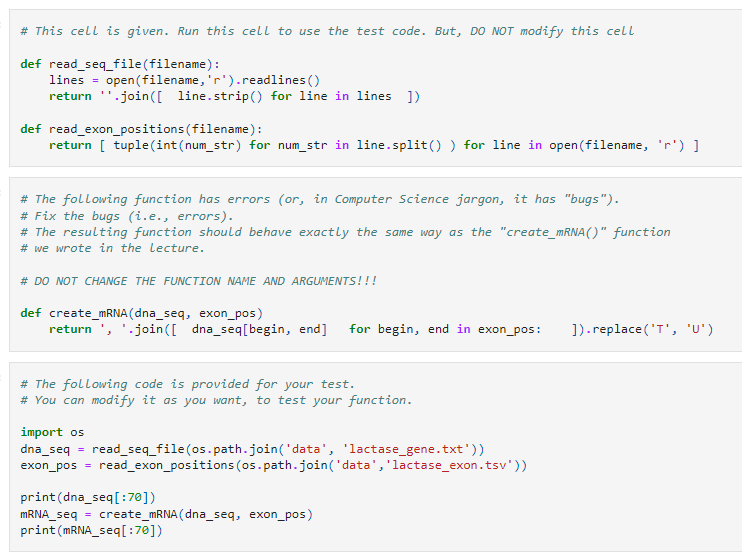
# This cell is given. Run this cell to use the test code. But, DO NOT modify this cell def read_seq_file(filename): lines = open(filename, 'r').readlines return ".join([ line.strip) for line in lines ]) def read_exon_positions (filename): return [ tuple(int (num_str) for num_str in line.split()) for line in open(filename, 'r')] # The following function has errors (or, in Computer Science jargon, it has "bugs"). # Fix the bugs (i.e., errors). # The resulting function should behave exactly the same way as the "create_mRNA)" function # we wrote in the lecture. # DO NOT CHANGE THE FUNCTION NAME AND ARGUMENTS!!! def create_mRNA (dna_seq, exon_pos) return', '.join([ dna_seq[begin, end] for begin, end in exon_pos: 1).replace('T', 'U') # The following code is provided for your test. # You can modify it as you want, to test your function. import os dna_seq read_seq_file(os.path.join('data', 'lactase_gene.txt')) exon_pos = read_exon_positions (os.path.join('data', 'lactase_exon.tsv')) print(dna_seq[:70]) mRNA_seq = create_mRNA( dna_seq, exon_pos) print(mRNA_seq[:70])
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


