Question: Using R Studio Programming - Type all your answers in R: Are the standard errors for B still reliable if the constant variance assumption doesn't
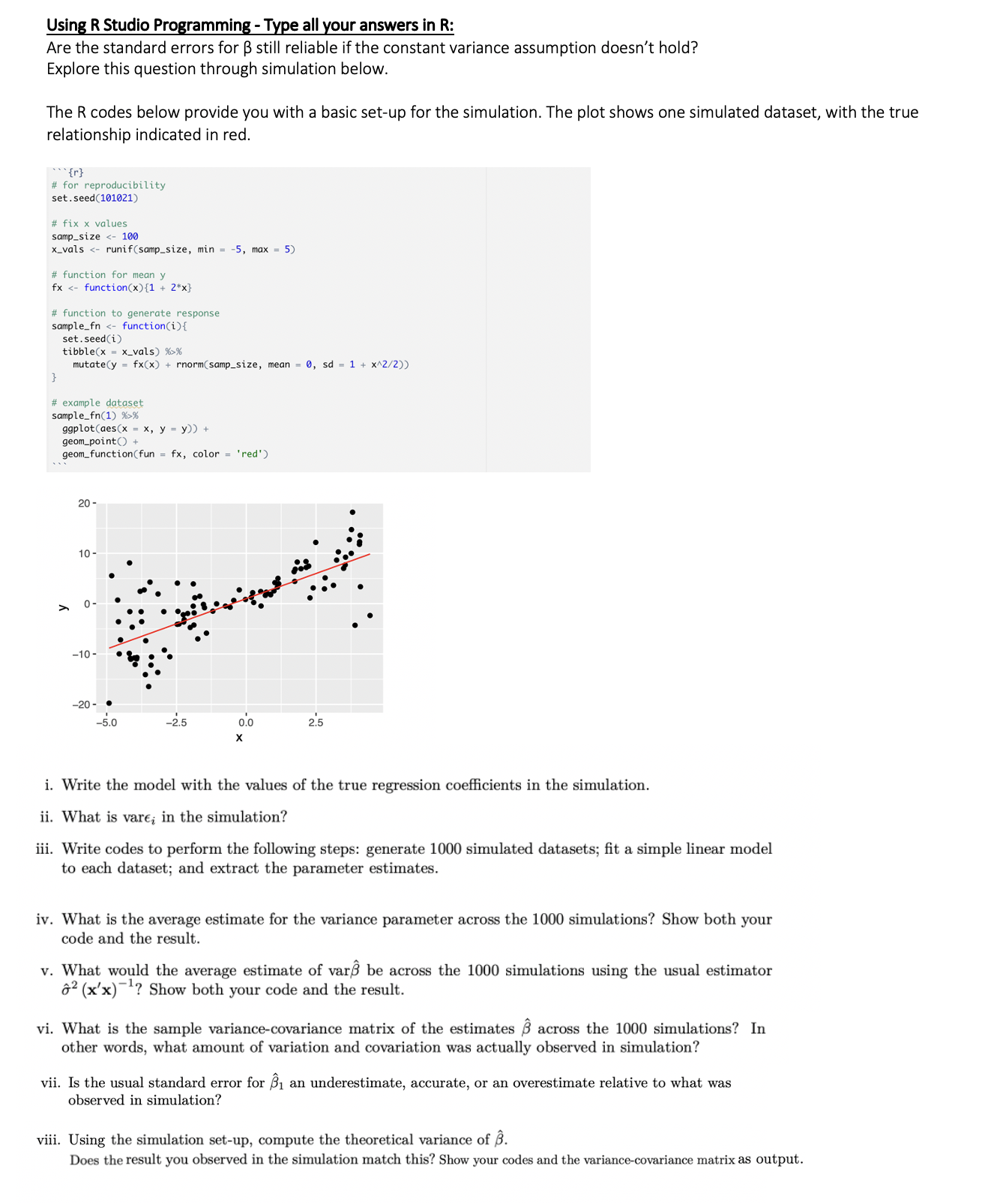
Using R Studio Programming - Type all your answers in R: Are the standard errors for B still reliable if the constant variance assumption doesn't hold? Explore this question through simulation below. The R codes below provide you with a basic set-up for the simulation. The plot shows one simulated dataset, with the true relationship indicated in red. "{r} for reproduciblety set. seed(101021) # fix x values samp_size % ggplot(aes(x = x, y = y)) + geom_point() + geom_function(fun = fx, color = 'red') 20 - 10 - -10 -20 - -5.0 -2.5 0.0 2.5 i. Write the model with the values of the true regression coefficients in the simulation. ii. What is vare; in the simulation? iii. Write codes to perform the following steps: generate 1000 simulated datasets; fit a simple linear model to each dataset; and extract the parameter estimates. iv. What is the average estimate for the variance parameter across the 1000 simulations? Show both your code and the result. v. What would the average estimate of varS be across the 1000 simulations using the usual estimator " (x'x)"? Show both your code and the result. vi. What is the sample variance-covariance matrix of the estimates B across the 1000 simulations? In other words, what amount of variation and covariation was actually observed in simulation? vii. Is the usual standard error for B1 an underestimate, accurate, or an overestimate relative to what was observed in simulation? viii. Using the simulation set-up, compute the theoretical variance of B. Does the result you observed in the simulation match this? Show your codes and the variance-covariance matrix as output
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


