Question: 23.46 DNA N-Gram Distribution (strings) This tool is provided by a third party. Your activity is always recorded, but you may need to refresh the
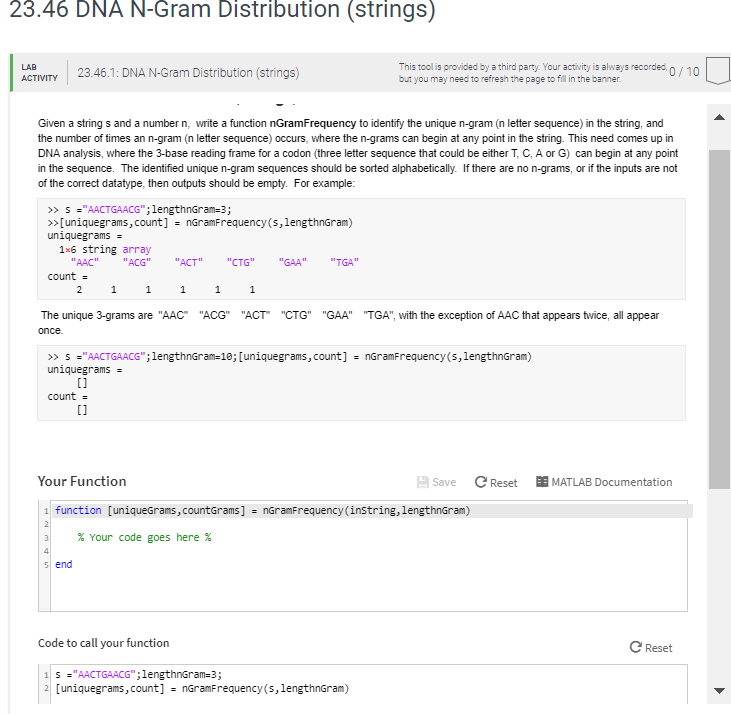
23.46 DNA N-Gram Distribution (strings) This tool is provided by a third party. Your activity is always recorded, but you may need to refresh the page to fl lin the banner LAB 23.46.1: DNA N-Gram Distribution (strings) 0/10 ACTIVITY Given a string s and a number n, write a function nGramFrequency to identify the unique n-gram (n letter sequence) in the string, and the number of times an n-gram (n letter sequence) occurs, where the n-grams can begi a any point in the string. This need comes up in DNA analysis, where the 3-base reading frame for a codon (three letter sequence that could be either T, C, A or G) can begin at any point in the sequence. The identified unique n-gram sequences should be sorted alphabetically. If there are no n-grams, or if the inputs are not of the cormect datatype, then outputs should be empty. For example >uniquegrams,count] nGramFrequency (s,lengthnGram) 1x6 string array "AAC"ACG"ACT""CTG" GAA"TGA count = The unique 3-grams are "AAC once "ACG" "ACT"CTG" "GAA" TGA", with the exception of AAC that appears twice, all appear >> s-"AACTGAACG" ; lengthnGram=10;[uniquegrams, count): nGran Frequency(s, lengthnGram) [l count = [l Your Function Save Reset E MATLAB Documentation 1 function [uniqueGrams, countGrams] nGramFrequency (instring,lengthnGram) % Your code goes here % s end Code to call your function C Reset 2 [uniquegrams,count] - nGramFrequency (s,lengthnGram)
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


