Question: help resolve the error in my code :import numpy as np # Given reaction mechanism ( coefficients are already corrected ) finput = [
help resolve the error in my code :import numpy as np
# Given reaction mechanism coefficients are already corrected
finput CH CH CHCH H H CHCH CH H
CH CH HCH CH CHCH CH CH CH
CH CH CH CHCH CH CH CH
reactions
for line in finput:
strippedline line.replace #
if in strippedline:
left, right strippedline.split
elif in strippedline:
left, right strippedline.split
else:
raise ValueErrorfInvalid delimiter in reaction: line
reactions.appendleftstrip right.strip
speciestmp
for left, right in reactions:
leftterms left.split
rightterms right.split
terms tstrip for t in lefttermststrip for t in rightterms
for term in terms:
tmp term.split
assert lentmp or lentmp 'terms r term r tmp rterms term, tmp
if lentmp:
speciestmpappendtmpstrip
else:
speciestmpappendtermstrip
speciesfiltered setspeciestmp
species listspeciesfiltered
# Initialize the stoichiometric matrix as zero
smtrx npzeroslenreactions lenspecies
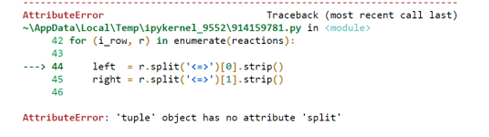
for irow, r in enumeratereactions:
left rsplitstrip
right rsplitstrip
leftterms left.split
leftterms tstrip for t in leftterms # inplace clean up
rightterms right.split
rightterms tstrip for t in rightterms # inplace clean up
for t in leftterms: # reactants
tmp tsplit # split stoichiometric coeff and species name
if lentmp: # stoich coeff and species name
coeff floattmpstrip
speciesmember tmpstrip
jcol species.indexspeciesmember # find id of species in the species list
assert smtrxirow,jcol
'duplicates not allowed rr: r r r
irow,rspeciesmember,smtrxirow,jcol
smtrxirow,jcol coeff
else: # only species name
speciesmember tmpstrip
jcol species.indexspeciesmember
assert smtrxirow,jcol
'duplicates not allowed rr: r r r
irow,rspeciesmember,smtrxirow,jcol
smtrxirow,jcol
for t in rightterms: # products
tmp tsplit
if lentmp:
coeff floattmpstrip
speciesmember tmpstrip
jcol species.indexspeciesmember
assert smtrxirow,jcol
'duplicates not allowed rr: r r r
irow,rspeciesmember,smtrxirow,jcol
smtrxirow,jcol coeff
else:
speciesmember tmpstrip
jcol species.indexspeciesmember
assert smtrxirow,jcol
'duplicates not allowed rr: r r r
irow,rspeciesmember,smtrxirow,jcol
smtrxirow,jcol
printspecies species
print# of species lenspecies
Step by Step Solution
There are 3 Steps involved in it
1 Expert Approved Answer
Step: 1 Unlock


Question Has Been Solved by an Expert!
Get step-by-step solutions from verified subject matter experts
Step: 2 Unlock
Step: 3 Unlock


