Question: in python please def count_dom_phenotype (genotypes): This function takes in a list of six nonnegative integers corresponding to the number of couples in a population

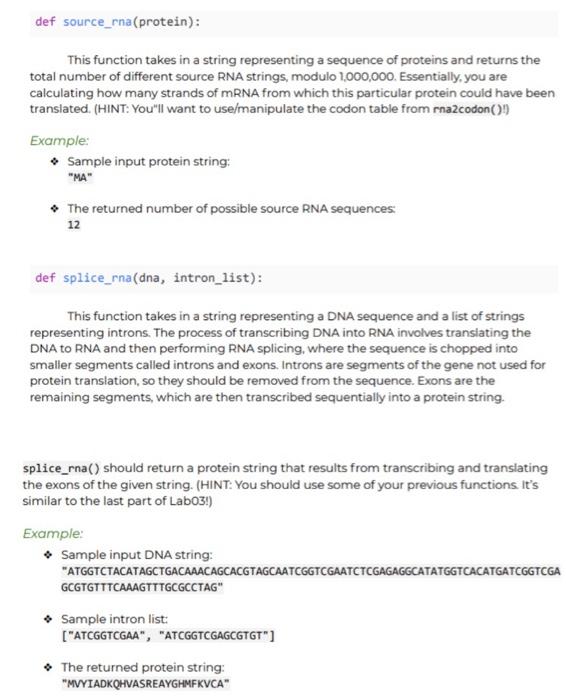
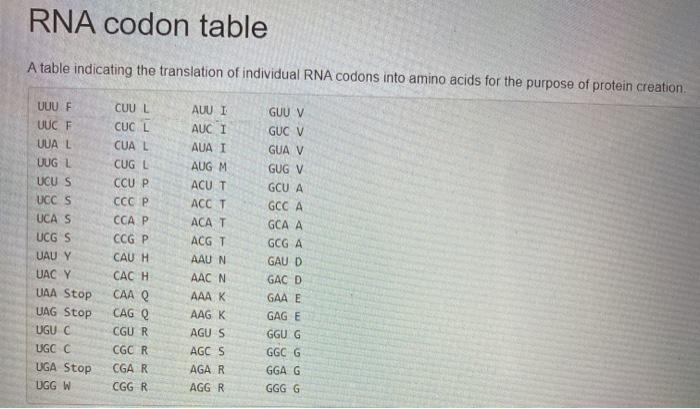
def count_dom_phenotype (genotypes): This function takes in a list of six nonnegative integers corresponding to the number of couples in a population possessing a specific genotype pairing, such as AA-AA (both homozygous dominant) or Aa-aa (heterozygous and homozygous recessive). The list of integers represent the number of couples having the following genotypes, in this order: 1. AA-AA 2. AA-Aa 3. AA-aa 4. Aa-Aa 5. Aa-aa 6. aa-aa The function should return the expected number of offspring displaying the dominant phenotype in the next generation, assuming that every couple has exactly two offspring. Example: Sample input list: [1, , , 1, 0, 1) The returned number of dominant phenotype offspring: 3.5 def source_rna (protein): This function takes in a string representing a sequence of proteins and returns the total number of different source RNA strings, modulo 1,000,000. Essentially, you are calculating how many strands of mRNA from which this particular protein could have been translated. (HINT: You'll want to use/manipulate the codon table from ma codon() Example: Sample input protein string: "MA" The returned number of possible source RNA sequences 12 def splice_rna(dna, intron_list): This function takes in a string representing a DNA sequence and a list of strings representing introns. The process of transcribing DNA into RNA involves translating the DNA to RNA and then performing RNA splicing, where the sequence is chopped into smaller segments called introns and exons. Introns are segments of the gene not used for protein translation, so they should be removed from the sequence. Exons are the remaining segments, which are then transcribed sequentially into a protein string. splice_rna() should return a protein string that results from transcribing and translating the exons of the given string. (HINT: You should use some of your previous functions It's similar to the last part of Labo3!) Example: Sample input DNA string: "ATGGTCTACATAGCTGACAAACAGCACGTAGCAATCGGTCGAATCTCGAGAGGCATATGGTCACATGATCGGTCGA GCGTGTTTCAAAGTTTGCGCCTAG" Sample intron list: ["ATCGGTCGAA", "ATCGGTCGAGCGTGT"] The returned protein string: "MVYIADKQHVASREAYGHMFKVCA" RNA codon table A table indicating the translation of individual RNA codons into amino acids for the purpose of protein creation UUU F UUC F UUA L UUGL UCU S UCC S UCA S UCG S UAU Y UACY UAA Stop UAG Stop UGU C UGC C UGA Stop UGG W CUU | CUCL CUAL CUG | CCUP CCCP CCA P CCG P CAUH CACH CAA Q CAGO CGUR CGC R CGA R CGGR AUU I AUCI AUA I AUGM ACUT ACCT ACGT AAUN AAC N AAA K AAG K AGUS AGCS AGAR AGGR GUU V GUC V GUA V GUG V GCU A GCC A GCA A GCG A GAU D GAC D GAA E GAGE GGU G GGC G GGA G GGGG
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


