Question: Table 11.3 Extinction coefficients for nucleic acids at 260 nm, in units of mL/(ug-cm) Double-stranded DNA 0.02 Single-stranded DNA 0.029 Double-stranded RNA 0.025 Oligonucleotides 0.04
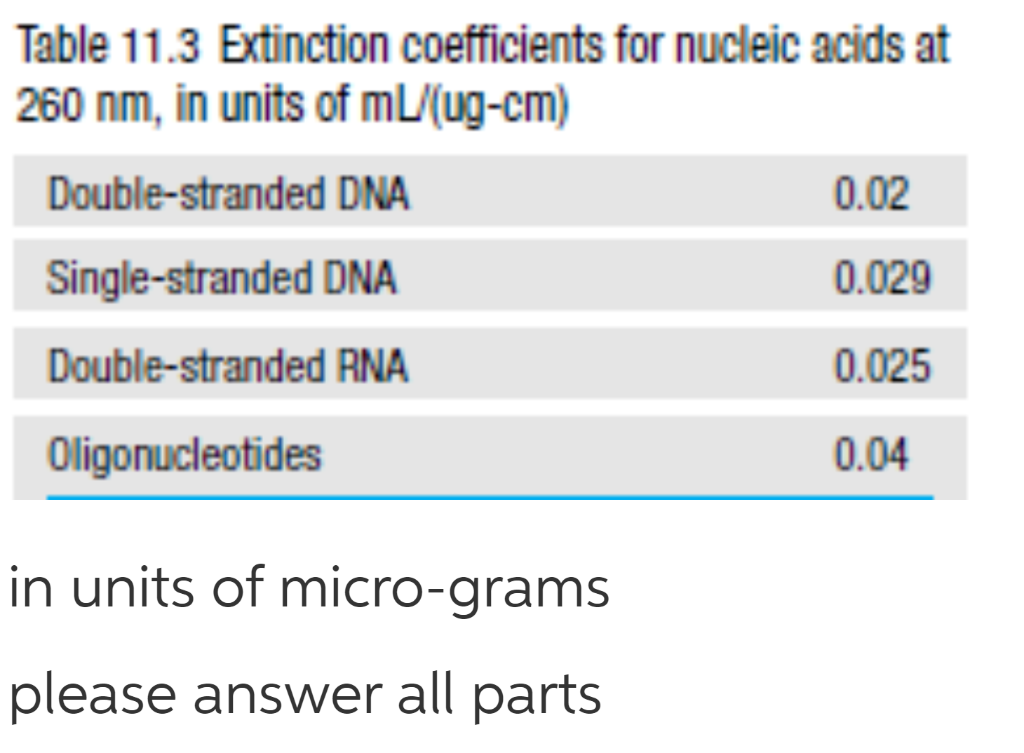

Table 11.3 Extinction coefficients for nucleic acids at 260 nm, in units of mL/(ug-cm) Double-stranded DNA 0.02 Single-stranded DNA 0.029 Double-stranded RNA 0.025 Oligonucleotides 0.04 in units of micro-grams please answer all parts In my lab, we use spectrophotometry to measure concentrations of DNA. My grad student adds 10 uL of double-stranded DNA to 990 uL of water, then takes an A260 with a 1 cm cuvette for a reading of 0.04. (a) Using table 11.3 in the text, what concentration of DNA is in the original (undiluted) sample? (b) To simplify this process, we have a machine called a Nanodrop, which uses only 1 uL of sample (so no dilution is necessary) and has a path length of 0.1 cm. What would the A260 reading on the Nanodrop be for the original (undiluted) sample? (c) The student needs to concentrate the DNA for the next application. She concentrates it to 1,000 ng/uL by removing 80% of the water. However, when she takes the reading on the Nanodrop, the machine tells her the concentration is only 900 ng/uL. Why might this be, given the expected A260 reading? (d) How could she get an accurate reading for DNA at this concentration using the Nanodrop
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


